Reading Data
Thomas W. Valente and George G. Vega Yon
Diffusion Network Object (diffnet)
netdiffuseR has its own class of objects:
diffnet.- Most of the package’s functions accept different types of graphs:
- Static:
matrix,dgCMatrix(from the Matrix pkg), - Dynamic:
list+dgCMatrix,array,diffnet
- Static:
But
diffnetis the class from which you get the most.From netdiffuseR’s perspective, network data comes in three classes:
- Raw R network data: Datasets with edgelist, attributes, survey data, etc.
- Already R data: already read into R using igraph, statnet, etc. (
igraph_to_diffnet,network_to_diffnet, etc.) - Graph files: DL, UCINET, pajek, etc. (
read_pajek,read_dl,read_ucinet, etc.)
In this presentation we will show focus on 1.
What is a diffnet object
A diffusion network, a.k.a. diffnet object, is a list that holds the following objects:
graph: Alistwith \(t\)dgCMatrixmatrices of size \(n\times n\),toa: An integer vector of length \(n\),adopt: A matrix of size \(n\times t\),cumadopt: A matrix of size \(n\times t\),vertex.static.attrs: Adata.frameof size \(n\times k\),vertex.dyn.attrs: A list with \(t\) dataframes of size \(n\times k\),graph.attrs: Currently ignored…, andmeta: A list with metadata about the object.
These are created using new_diffnet (or its wrappers).
Basic Diffusion Network
To create
diffnetobjects we only need a network and times of adoption:set.seed(9) # Network net <- rgraph_ws(500, 4, .2) # Times of adoption toa <- sample(c(NA, 2010:2014), 500, TRUE) diffnet_static <- as_diffnet(net, toa)# Warning in new_diffnet(graph, ...): -graph- is static and will be recycled # (see ?new_diffnet).summary(diffnet_static)# Diffusion network summary statistics # Name : Diffusion Network # Behavior : Unspecified # ----------------------------------------------------------------------------- # Period Adopters Cum Adopt. (%) Hazard Rate Density Moran's I (sd) # -------- ---------- ---------------- ------------- --------- ---------------- # 2010 94 94 (0.19) - 0.01 -0.00 (0.00) # 2011 67 161 (0.32) 0.17 0.01 -0.00 (0.00) # 2012 71 232 (0.46) 0.21 0.01 -0.00 (0.00) # 2013 93 325 (0.65) 0.35 0.01 -0.00 (0.00) # 2014 82 407 (0.81) 0.47 0.01 -0.00 (0.00) # ----------------------------------------------------------------------------- # Left censoring : 0.19 (94) # Right centoring : 0.19 (93) # # of nodes : 500 # # Moran's I was computed on contemporaneous autocorrelation using 1/geodesic # values. Significane levels *** <= .01, ** <= .05, * <= .1.
Static survey
netdiffuseR can also read survey (nomination) data:
data("fakesurvey") fakesurvey# id toa group net1 net2 net3 age gender # 1 1 1 1 NA NA NA 30 M # 2 2 5 1 3 1 NA 35 F # 3 3 5 1 NA 2 NA 31 F # 4 4 3 1 6 5 NA 30 M # 5 5 2 1 4 4 3 40 F # 6 1 4 2 3 4 8 29 F # 7 2 3 2 3 NA NA 35 M # 8 5 3 2 10 1 NA 50 M # 9 10 NA 2 5 1 NA 19 F # note # 1 No nominations # 2 Nothing weird # 3 Only nominates in net2 # 4 Nominates someone who wasn't interview # 5 Nominates 4 two times # 6 Only nominates outsiders # 7 Isolated # 8 Nothing weird # 9 Non-adopterIn group one 4 nominates id 6, who does not show in the data, and in group two 1 nominates 3, 4, and 8, also individuals who don’t show up in the survey.
d1 <- survey_to_diffnet( dat = fakesurvey, # Dataset idvar = "id", # The name of the idvar netvars = c("net1", "net2", "net3"), # Name of the nomination variables groupvar = "group", # Group variable (if any) toavar = "toa" # Name of the time of adoption variable ); d1# Dynamic network of class -diffnet- # Name : Diffusion Network # Behavior : Unspecified # # of nodes : 9 (101, 102, 103, 104, 105, 201, 202, 205, ...) # # of time periods : 5 (1 - 5) # Type : directed # Final prevalence : 0.89 # Static attributes : group, net1, net2, net3, age, gender, note (7) # Dynamic attributes : -If you want to include those, you can use the option
no.unsurveyedd2 <- survey_to_diffnet( dat = fakesurvey, idvar = "id", netvars = c("net1", "net2", "net3"), groupvar = "group", toavar = "toa", no.unsurveyed = FALSE ); d2# Dynamic network of class -diffnet- # Name : Diffusion Network # Behavior : Unspecified # # of nodes : 13 (101, 102, 103, 104, 105, 106, 201, 202, ...) # # of time periods : 5 (1 - 5) # Type : directed # Final prevalence : 0.62 # Static attributes : group, net1, net2, net3, age, gender, note (7) # Dynamic attributes : -We can also check the difference
d2 - d1# Dynamic network of class -diffnet- # Name : Diffusion Network # Behavior : Unspecified # # of nodes : 4 (106, 203, 204, 208) # # of time periods : 5 (1 - 5) # Type : directed # Final prevalence : 0.00 # Static attributes : group, net1, net2, net3, age, gender, note (7) # Dynamic attributes : -rownames(d2 - d1)# [1] "106" "203" "204" "208"
Dynamic survey
data("fakesurveyDyn")
fakesurveyDyn# id toa group net1 net2 net3 age gender
# 1 1 1991 1 NA NA NA 30 M
# 2 2 1990 1 3 1 NA 35 F
# 3 3 1991 1 NA 2 NA 31 F
# 4 4 1990 1 6 5 NA 30 M
# 5 5 1991 1 4 4 3 40 F
# 6 1 1991 2 3 4 8 29 F
# 7 2 1990 2 3 NA NA 35 M
# 8 5 1990 2 10 1 NA 50 M
# 9 10 1990 2 5 1 NA 19 F
# 10 1 1991 1 NA NA NA 31 M
# 11 2 1990 1 3 1 NA 36 F
# 12 3 1991 1 NA 2 NA 32 F
# 13 4 1990 1 6 5 NA 31 M
# 14 5 1991 1 4 4 3 41 F
# 15 1 1991 2 3 4 8 30 F
# 16 2 1990 2 1 NA NA 36 M
# 17 5 1990 2 10 1 NA 51 M
# 18 10 1990 2 5 1 NA 20 F
# note time
# 1 First wave: No nominations 1990
# 2 First wave: Nothing weird 1990
# 3 First wave: Only nominates in net2 1990
# 4 First wave: Nominates someone who wasn't interview 1990
# 5 First wave: Nominates 4 two times 1990
# 6 First wave: Only nominates outsiders 1990
# 7 First wave: Isolated 1990
# 8 First wave: Nothing weird 1990
# 9 First wave: Non-adopter 1990
# 10 Second wave: No nominations 1991
# 11 Second wave: Nothing weird 1991
# 12 Second wave: Only nominates in net2 1991
# 13 Second wave: Nominates someone who wasn't interview 1991
# 14 Second wave: Nominates 4 two times 1991
# 15 Second wave: Only nominates outsiders 1991
# 16 Second wave: Now is not isolated! 1991
# 17 Second wave: Nothing weird 1991
# 18 Second wave: Non-adopter 1991diffnet_dynsurvey <- survey_to_diffnet(
dat = fakesurveyDyn,
idvar = "id",
netvars = c("net1", "net2", "net3"),
groupvar = "group",
toavar = "toa",
timevar = "time"
)# Warning in check_var_class_and_coerce(x, dat, c("numeric", "integer"),
# "integer", : Coercing -net1- into integer.# Warning in check_var_class_and_coerce(x, dat, c("numeric", "integer"),
# "integer", : Coercing -time- into integer.plot_diffnet(diffnet_dynsurvey)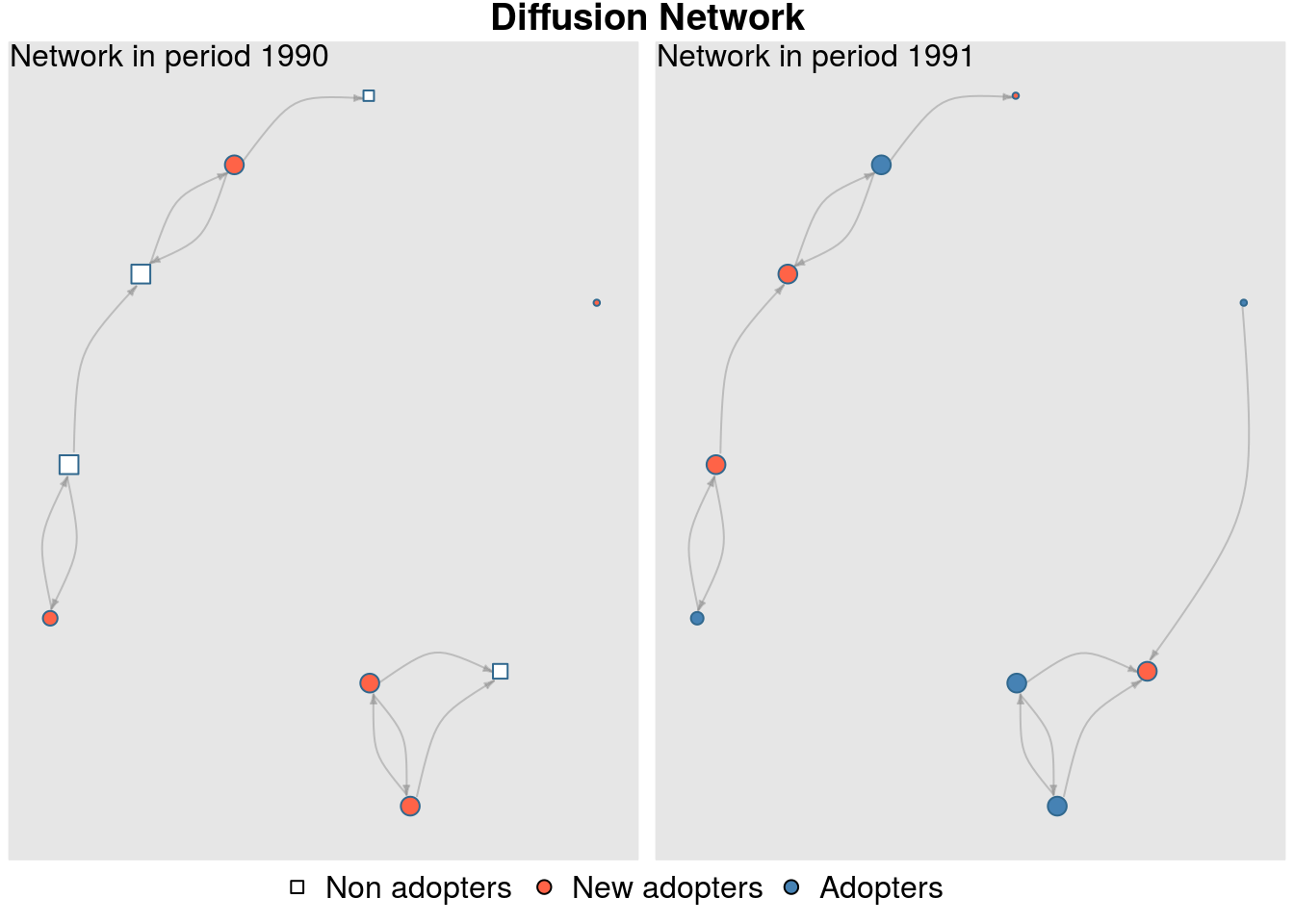
Other formats
The package also supports working with other network formats.
Besides of
.net(Pajek), andml(UCINET), netdiffuseR can actually convert between classes:igraph,network, andnetworkDynamic.data("medInnovationsDiffNet") dn_ig <- diffnet_to_igraph(medInnovationsDiffNet) # dn_ig # For some issue with lazy eval, knitr won't print this dn_net <- diffnet_to_network(medInnovationsDiffNet) dn_net[[1]]# Network attributes: # vertices = 125 # directed = TRUE # hyper = FALSE # loops = FALSE # multiple = FALSE # bipartite = FALSE # name = Medical Innovation # behavior = Adoption of Tetracycline # total edges= 294 # missing edges= 0 # non-missing edges= 294 # # Vertex attribute names: # ado adopt attend belief catbak city club coll commun ctl date detail detail2 dichot drug expect free friends here home house info journ journ2 length meet most net1_1 net1_2 net1_3 net2_1 net2_2 net2_3 net3_1 net3_2 net3_3 nojourn nonpoor office origid paadico perc position presc proage proage2 proximty recall recon reltend science social sourinfo special study tend thresh toa vertex.names young # # No edge attributesdn_ndy <- diffnet_to_networkDynamic(medInnovationsDiffNet)# Argument base.net not specified, using first element of network.list instead # Created net.obs.period to describe network # Network observation period info: # Number of observation spells: 1 # Maximal time range observed: 1 until 18 # Temporal mode: discrete # Time unit: step # Suggested time increment: 1dn_ndy# NetworkDynamic properties: # distinct change times: 18 # maximal time range: 1 until 18 # # Includes optional net.obs.period attribute: # Network observation period info: # Number of observation spells: 1 # Maximal time range observed: 1 until 18 # Temporal mode: discrete # Time unit: step # Suggested time increment: 1 # # Network attributes: # vertices = 125 # directed = TRUE # hyper = FALSE # loops = FALSE # multiple = FALSE # bipartite = FALSE # behavior = Adoption of Tetracycline # name = Medical Innovation # net.obs.period: (not shown) # total edges= 294 # missing edges= 0 # non-missing edges= 294 # # Vertex attribute names: # active ado adopt attend belief catbak city club coll commun ctl date detail detail2 dichot drug expect free friends here home house info journ journ2 length meet most net1_1 net1_2 net1_3 net2_1 net2_2 net2_3 net3_1 net3_2 net3_3 nojourn nonpoor office origid paadico perc position presc proage proage2 proximty recall recon reltend science social sourinfo special study tend thresh toa vertex.names young # # Edge attribute names: # activeFirst two examples it creates a list of objects, the later actually creates a single object
networkDynamic_to_diffnet(dn_ndy, toavar = "toa")# Dynamic network of class -diffnet- # Name : Medical Innovation # Behavior : Adoption of Tetracycline # # of nodes : 125 (1001, 1002, 1003, 1004, 1005, 1006, 1007, 1008, ...) # # of time periods : 18 (1 - 18) # Type : directed # Final prevalence : 1.00 # Static attributes : - # Dynamic attributes : ado, adopt, attend, belief, catbak, city, club, co... (59)
Problems
- Using the rda file read.rda, read in the edgelist
net_edgelistand the adjacency matrixnet_listas a diffnet objects. In both cases you should use the data.frameXwhich has the time of adoption variable. (solution script)
Center for Applied Network Analysis (CANA)