A simple simulation example
George G Vega Yon
February 25, 2019
Source:vignettes/rdiffnets.Rmd
rdiffnets.RmdStep 2: Random network (for input)
set.seed(8826)
# Simulating a small world (10, 3) with pr = .3
net <- rgraph_ws(50, 3, .3)
# A bit more of rewiring
net <- rewire_graph(net, p=3, both.ends = TRUE)## Warning in rewire_graph(net, p = 3, both.ends = TRUE): The option -copy.first-
## is set to TRUE. In this case, the first graph will be treated as a baseline,
## and thus, networks after T=1 will be replaced with T-1.Taking a look at it using sna

# Random diffusion with a fixed threshold of 1, simulating 5 time points
mydiffnet <- rdiffnet(
seed.graph = net, # The network we just created
threshold.dist = 1, # Fixed threshold of 1
t = 5, # 5 time points
rewire = FALSE, # No rewire (defaults TRUE)
exposure.args = list(normalized=FALSE), # Exposure to be computed unnormalized
# so we use counts instead
seed.nodes = "random" # Random set of initial adopters
)
# Looking at the diffusion process
plot_diffnet(mydiffnet)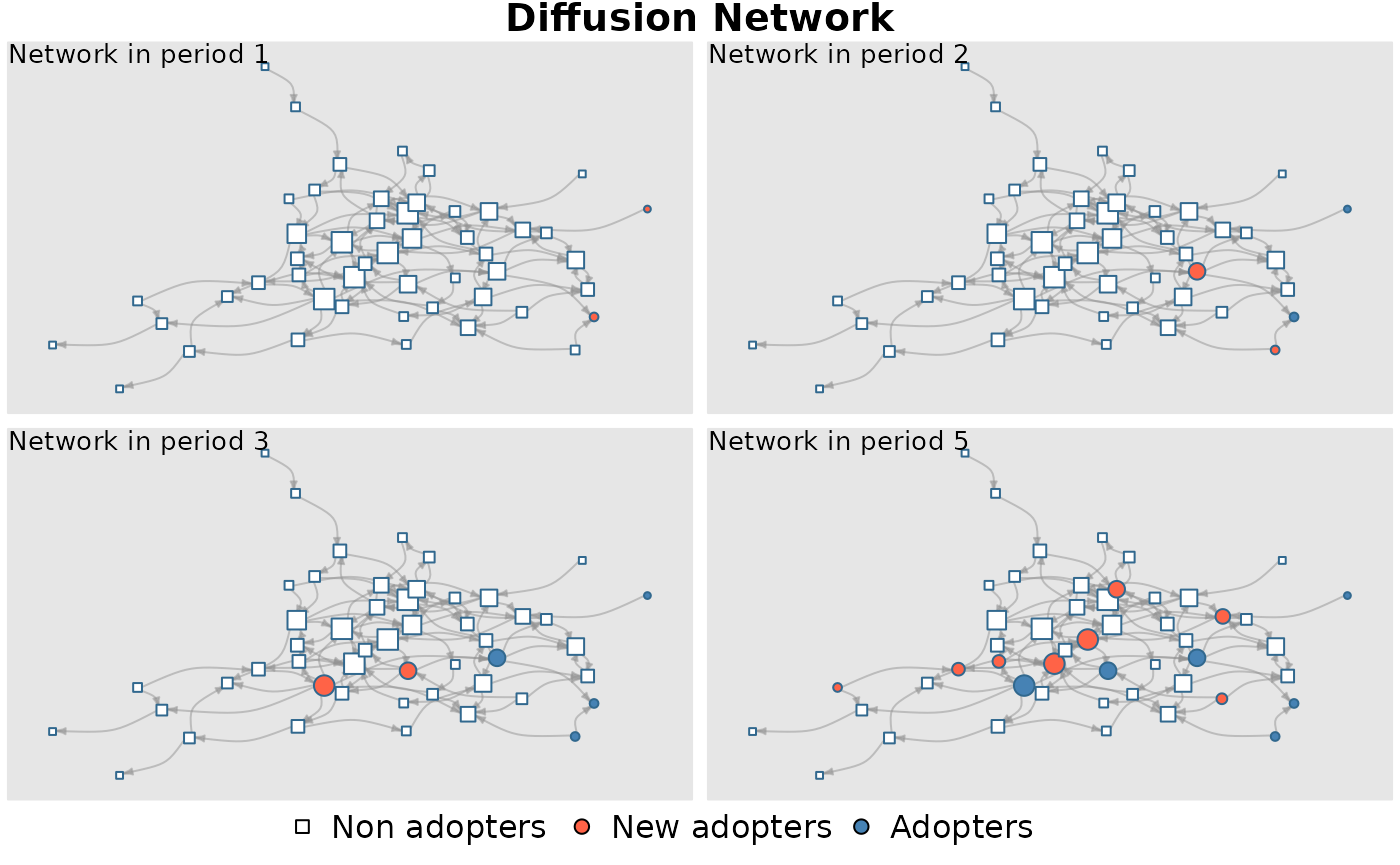
# Some summary stats
summary(mydiffnet)## Diffusion network summary statistics
## Name : A diffusion network
## Behavior : Random contagion
## -----------------------------------------------------------------------------
## Period Adopters Cum Adopt. (%) Hazard Rate Density Moran's I (sd)
## -------- ---------- ---------------- ------------- --------- ----------------
## 1 2 2 (0.04) - 0.04 0.00 (0.03)
## 2 2 4 (0.08) 0.04 0.04 0.15 (0.03) ***
## 3 2 6 (0.12) 0.04 0.04 0.10 (0.03) ***
## 4 3 9 (0.18) 0.07 0.04 0.08 (0.03) ***
## 5 5 14 (0.28) 0.12 0.04 0.07 (0.04) ***
## -----------------------------------------------------------------------------
## Left censoring : 0.04 (2)
## Right centoring : 0.72 (36)
## # of nodes : 50
##
## Moran's I was computed on contemporaneous autocorrelation using 1/geodesic
## values. Significane levels *** <= .01, ** <= .05, * <= .1.We can actually go further and run multiple simulations instead so that we can get a confidence interval in the proportion of adopters
set.seed(871)
mydiffnet <- rdiffnet_multiple(
statistic = function(n) cumulative_adopt_count(n)["prop",],
R = 1000,
stop.no.diff = FALSE, # This option allows us to continue
# The simulation process, even if there
# is no adoption.
seed.graph = net, # The network we just created
threshold.dist = 1, # Fixed threshold of 1
t = 5, # 5 time points
rewire = FALSE, # No rewire (defaults TRUE)
exposure.args = list(normalized=FALSE), # Exposure to be computed unnormalized
# so we use counts instead
seed.nodes = "random" # Random set of initial adopters
)
# Looking at the diffusion process
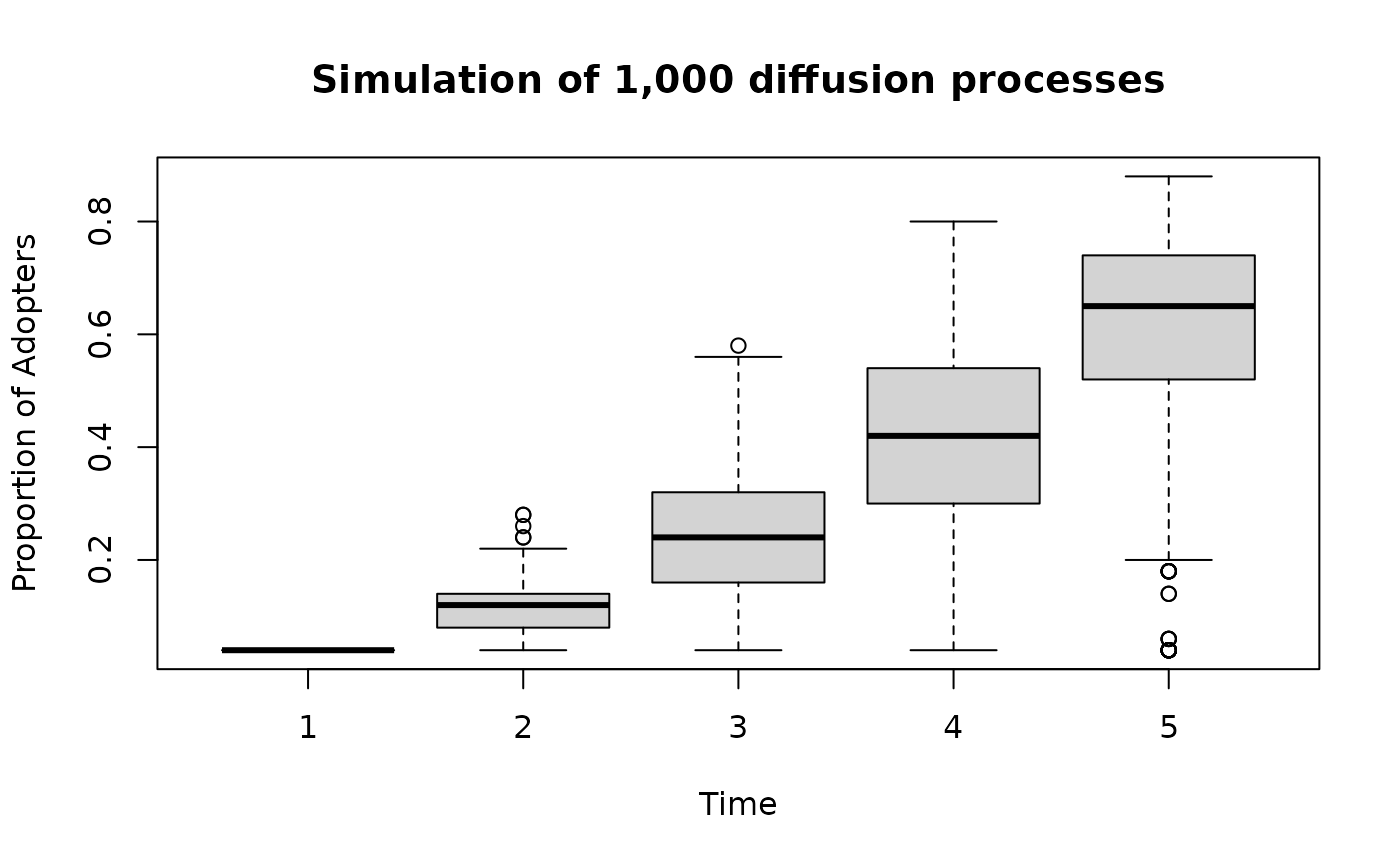
boxplot(
t(mydiffnet),
xlab = "Time",
ylab = "Proportion of Adopters",
main = "Simulation of 1,000 diffusion processes"
)