Simulating diffusion networks: Using the
rdiffnet function
George G. Vega Yon
November 17, 2015
Source:vignettes/introduction-to-netdiffuser.Rmd
introduction-to-netdiffuser.RmdSimulating diffusion networks: Using the rdiffnet
function
In this example we compare 3 different simulations that use the same
baseline (seed) network, a scale-free generated via
rgraph_ba (Barabasi-Albert) with parameter m=4
(number of new ties that each added node includes in the graph). The
only difference between the three simulations is that we use a different
set of seed adopters, “random”, “central” and “marginal”. All three
cases start with 5% of the network having adopted the innovation.
##
## Attaching package: 'netdiffuseR'## The following object is masked from 'package:base':
##
## %*%
s <- 11532
set.seed(s)
diffnet_ran <- rdiffnet(200, 20, "random", seed.p.adopt = .1,
seed.graph = "small-world",
rgraph.args = list(undirected=FALSE, k=4, p=.5),
threshold.dist = function(x) 0.3)## Warning in (function (graph, p, algorithm = "endpoints", both.ends = FALSE, :
## The option -copy.first- is set to TRUE. In this case, the first graph will be
## treated as a baseline, and thus, networks after T=1 will be replaced with T-1.
set.seed(s)
diffnet_cen <- rdiffnet(200, 20, "central", seed.p.adopt = .1,
seed.graph = "small-world",
rgraph.args = list(undirected=FALSE, k=4, p=.5),
threshold.dist = function(x) 0.3)## Warning in (function (graph, p, algorithm = "endpoints", both.ends = FALSE, :
## The option -copy.first- is set to TRUE. In this case, the first graph will be
## treated as a baseline, and thus, networks after T=1 will be replaced with T-1.
set.seed(s)
diffnet_mar <- rdiffnet(200, 20, "marginal", seed.p.adopt = .1,
seed.graph = "small-world",
rgraph.args = list(undirected=FALSE, k=4, p=.5),
threshold.dist = function(x) 0.3)## Warning in (function (graph, p, algorithm = "endpoints", both.ends = FALSE, :
## The option -copy.first- is set to TRUE. In this case, the first graph will be
## treated as a baseline, and thus, networks after T=1 will be replaced with T-1.Furthermore, we can take a more detail view of what’s going on in
each network using the summary method. For example, lets
take a look at the marginal network:
summary(diffnet_mar)## Diffusion network summary statistics
## Name : A diffusion network
## Behavior : Random contagion
## -----------------------------------------------------------------------------
## Period Adopters Cum Adopt. (%) Hazard Rate Density Moran's I (sd)
## -------- ---------- ---------------- ------------- --------- ----------------
## 1 20 20 (0.10) - 0.02 -0.01 (0.00)
## 2 2 22 (0.11) 0.01 0.02 -0.00 (0.00)
## 3 2 24 (0.12) 0.01 0.02 -0.00 (0.00)
## 4 1 25 (0.12) 0.01 0.02 -0.00 (0.00)
## 5 1 26 (0.13) 0.01 0.02 -0.00 (0.00) *
## 6 4 30 (0.15) 0.02 0.02 0.00 (0.00) ***
## 7 1 31 (0.15) 0.01 0.02 0.00 (0.00) ***
## 8 5 36 (0.18) 0.03 0.02 0.00 (0.00) ***
## 9 4 40 (0.20) 0.02 0.02 0.01 (0.00) ***
## 10 5 45 (0.23) 0.03 0.02 0.01 (0.00) ***
## 11 13 58 (0.29) 0.08 0.02 0.01 (0.00) ***
## 12 15 73 (0.36) 0.11 0.02 0.02 (0.00) ***
## 13 21 94 (0.47) 0.17 0.02 0.02 (0.00) ***
## 14 36 130 (0.65) 0.34 0.02 0.01 (0.00) ***
## 15 46 176 (0.88) 0.66 0.02 0.00 (0.00) ***
## 16 23 199 (0.99) 0.96 0.02 -0.01 (0.00)
## 17 1 200 (1.00) 1.00 0.02 -
## 18 0 200 (1.00) 0.00 0.02 -
## 19 0 200 (1.00) 0.00 0.02 -
## 20 0 200 (1.00) 0.00 0.02 -
## -----------------------------------------------------------------------------
## Left censoring : 0.10 (20)
## Right centoring : 0.00 (0)
## # of nodes : 200
##
## Moran's I was computed on contemporaneous autocorrelation using 1/geodesic
## values. Significane levels *** <= .01, ** <= .05, * <= .1.At a first look, printing the networks, we can see that they differ in the number of adopters, as the adoption rate shows:
diffnet_ran; diffnet_cen; diffnet_mar## Dynamic network of class -diffnet-
## Name : A diffusion network
## Behavior : Random contagion
## # of nodes : 200 (1, 2, 3, 4, 5, 6, 7, 8, ...)
## # of time periods : 20 (1 - 20)
## Type : directed
## Final prevalence : 1.00
## Static attributes : real_threshold (1)
## Dynamic attributes : -## Dynamic network of class -diffnet-
## Name : A diffusion network
## Behavior : Random contagion
## # of nodes : 200 (1, 2, 3, 4, 5, 6, 7, 8, ...)
## # of time periods : 20 (1 - 20)
## Type : directed
## Final prevalence : 1.00
## Static attributes : real_threshold (1)
## Dynamic attributes : -## Dynamic network of class -diffnet-
## Name : A diffusion network
## Behavior : Random contagion
## # of nodes : 200 (1, 2, 3, 4, 5, 6, 7, 8, ...)
## # of time periods : 20 (1 - 20)
## Type : directed
## Final prevalence : 1.00
## Static attributes : real_threshold (1)
## Dynamic attributes : -So, as expected, the network that used central nodes as first adopters is the one that reached the highest adoption rate, 0.95; meanwhile the network that used marginal nodes as seed has the lowest adoption rate, 0.56. Lets compare the set of initial adopters graphically
cols <- c("lightblue","green", "blue")
oldpar <- par(no.readonly = TRUE)
par(mfcol=c(1,3), mai = c(0, 0, 1, 0), mar = rep(1, 4) + 0.1)
set.seed(s);plot(diffnet_ran, main="Random seed")## Warning in sprintf(main, x$meta$pers[t]): one argument not used by format
## 'Random seed'## Warning in sprintf(main, x$meta$pers[t]): one argument not used by format
## 'Central seed'## Warning in sprintf(main, x$meta$pers[t]): one argument not used by format
## 'Marginal seed'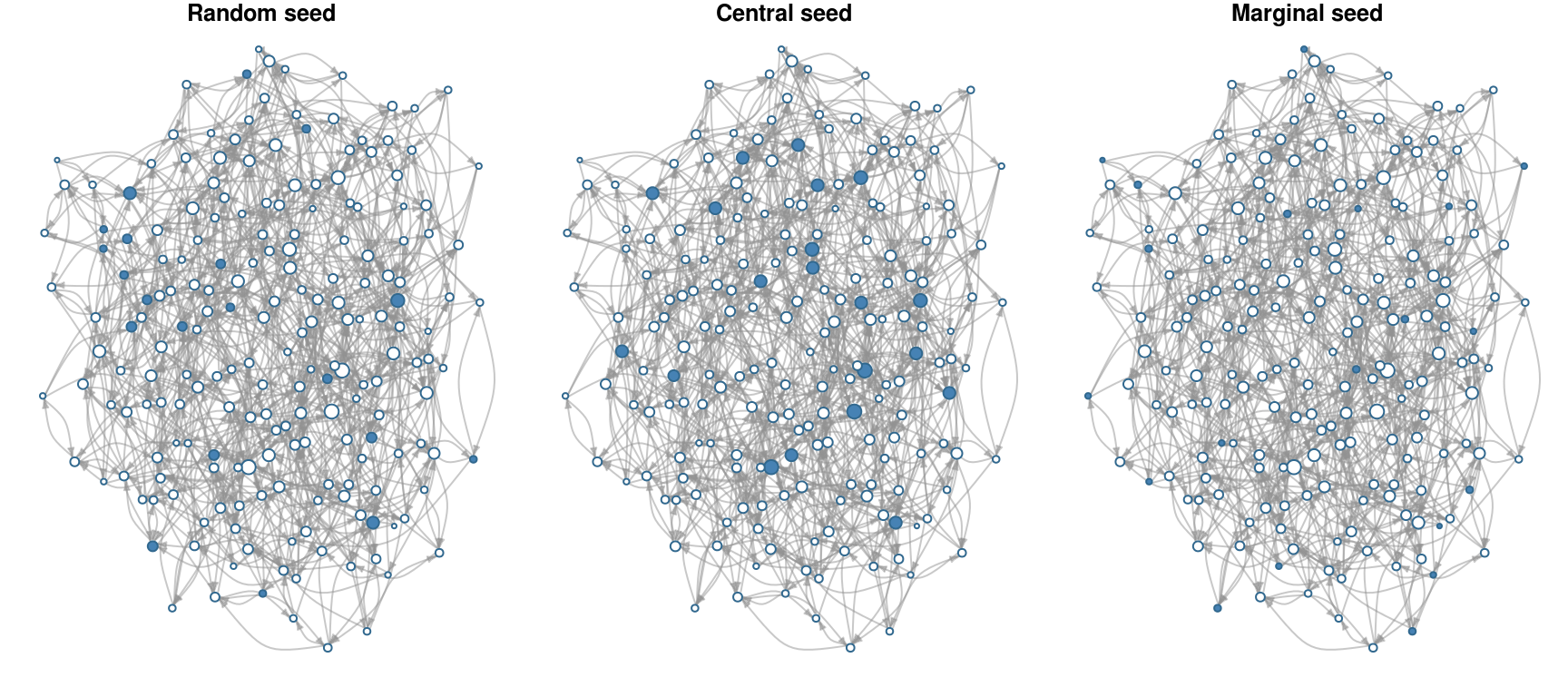
par(oldpar)An interesting way of visualizing the diffusion process is using the
plot_diffnet function. In this case, instead of plotting
all the 20 periods (networks), we only focus on a subset (henceforth we
use the slices argument).
plot_diffnet(diffnet_ran, slices = c(1,4,8,12,16,20), layout=coords)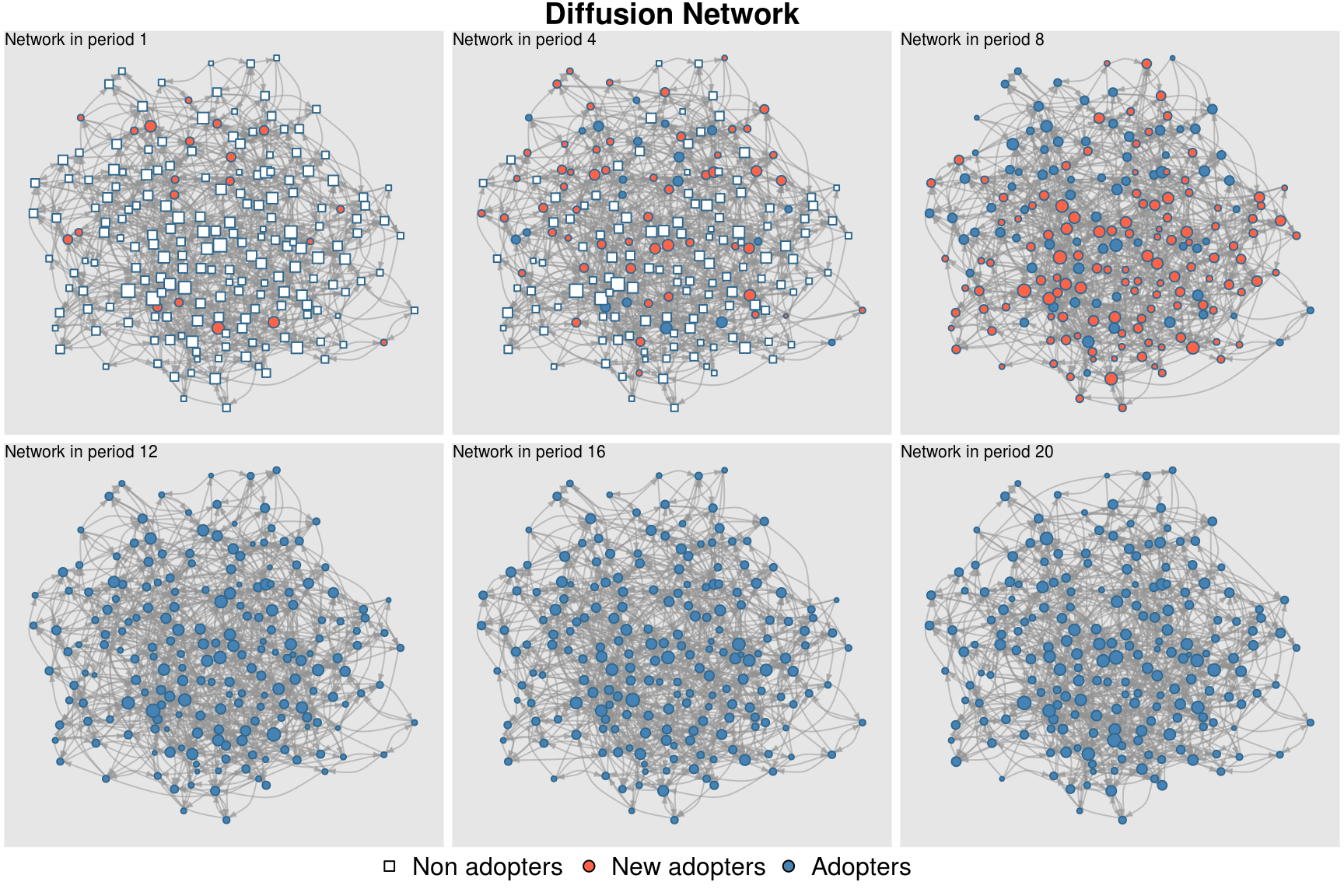
Diffusion process
An easy way to compare these three networks is by checking the
cumulative adoption counts, in particular, the proportion. Using the
function plot_adopters we can achieve our goal
plot_adopters(diffnet_ran, bg = cols[1], include.legend = FALSE, what="cumadopt")
plot_adopters(diffnet_cen, bg = cols[2], add=TRUE, what="cumadopt")
plot_adopters(diffnet_mar, bg = cols[3], add=TRUE, what="cumadopt")
legend("topleft", bty="n",
legend = c("Random","Central", "Marginal"),
fill=cols)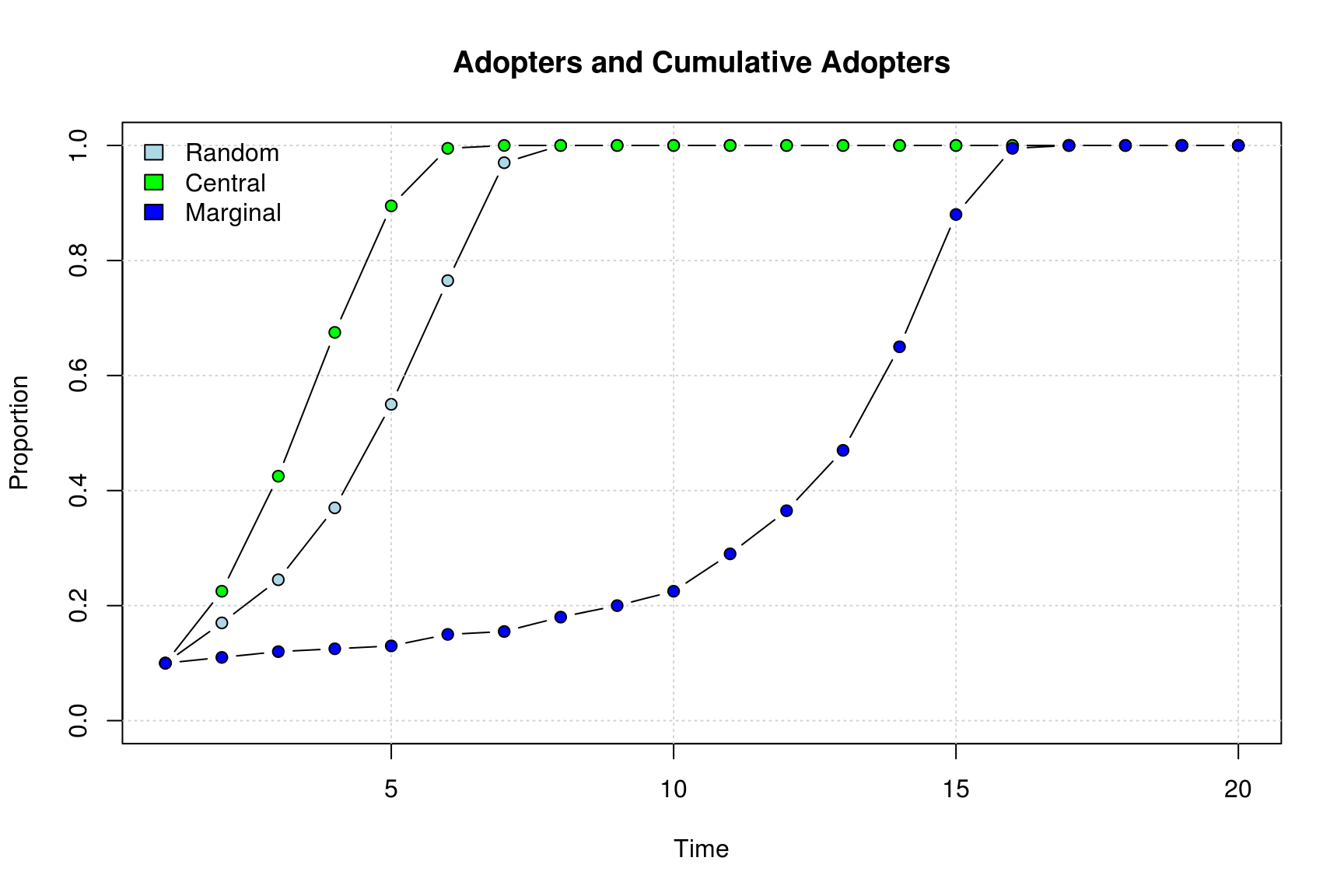
Comparing hazard rates we can do the following
plot_hazard(diffnet_ran, ylim=c(0,1), bg=cols[1])
plot_hazard(diffnet_cen, add=TRUE, bg=cols[2])
plot_hazard(diffnet_mar, add=TRUE, bg=cols[3])
legend("topleft", bty="n",
legend = c("Random","Central", "Marginal"),
fill=cols)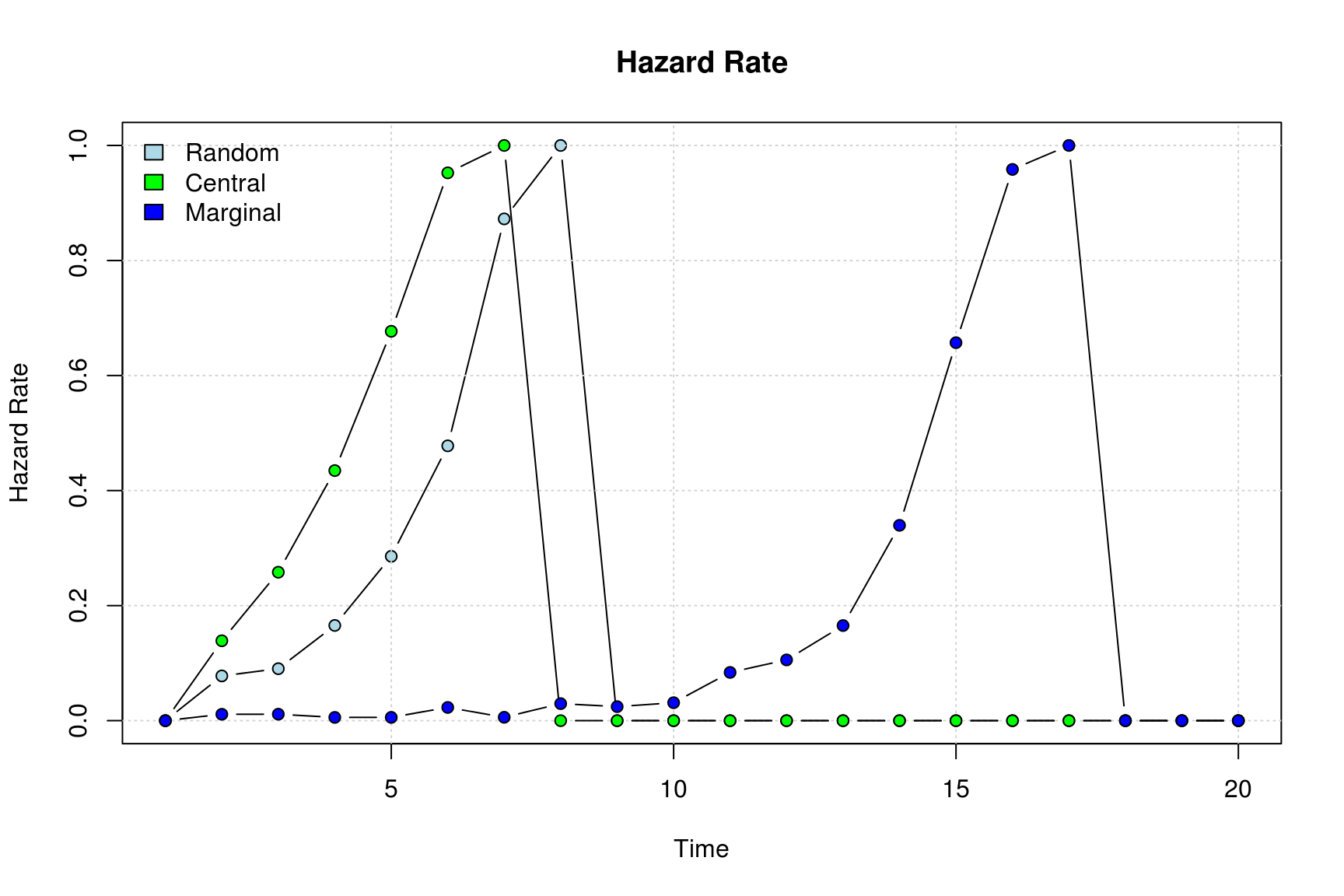
Furthermore, we can calculate infectiousness and susceptibility on each network and see whether both are correlated in each one of the processess.
plot_infectsuscep(diffnet_ran, bins=15, K=3,
main = "Distribution of Infectiousness and\nSusceptibility (Random)")## Warning in plot_infectsuscep.list(graph$graph, graph$toa, t0, normalize, : When
## applying logscale some observations are missing.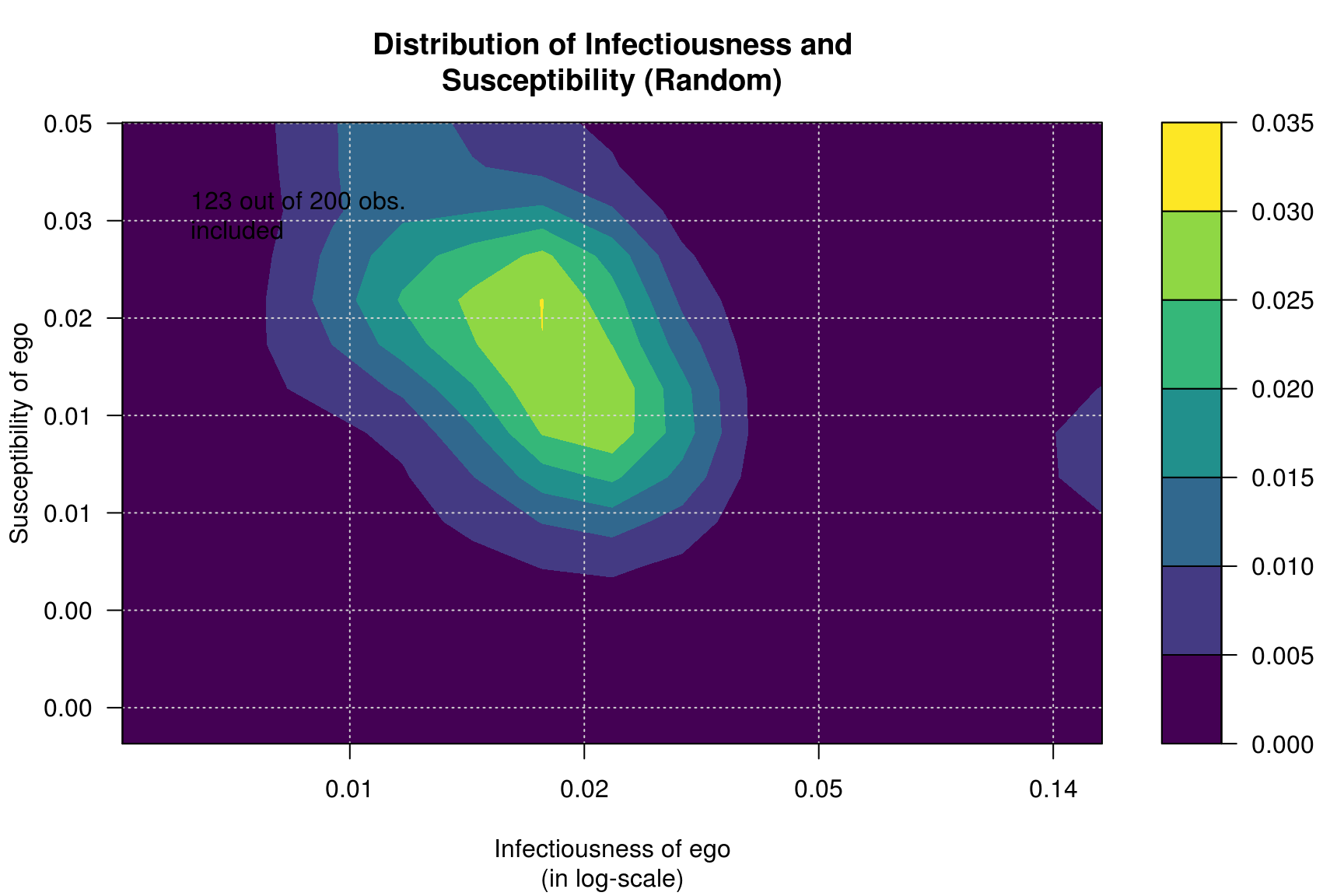
plot_infectsuscep(diffnet_cen, bins=15, K=3,
main = "Distribution of Infectiousness and\nSusceptibility (Central)")## Warning in plot_infectsuscep.list(graph$graph, graph$toa, t0, normalize, : When
## applying logscale some observations are missing.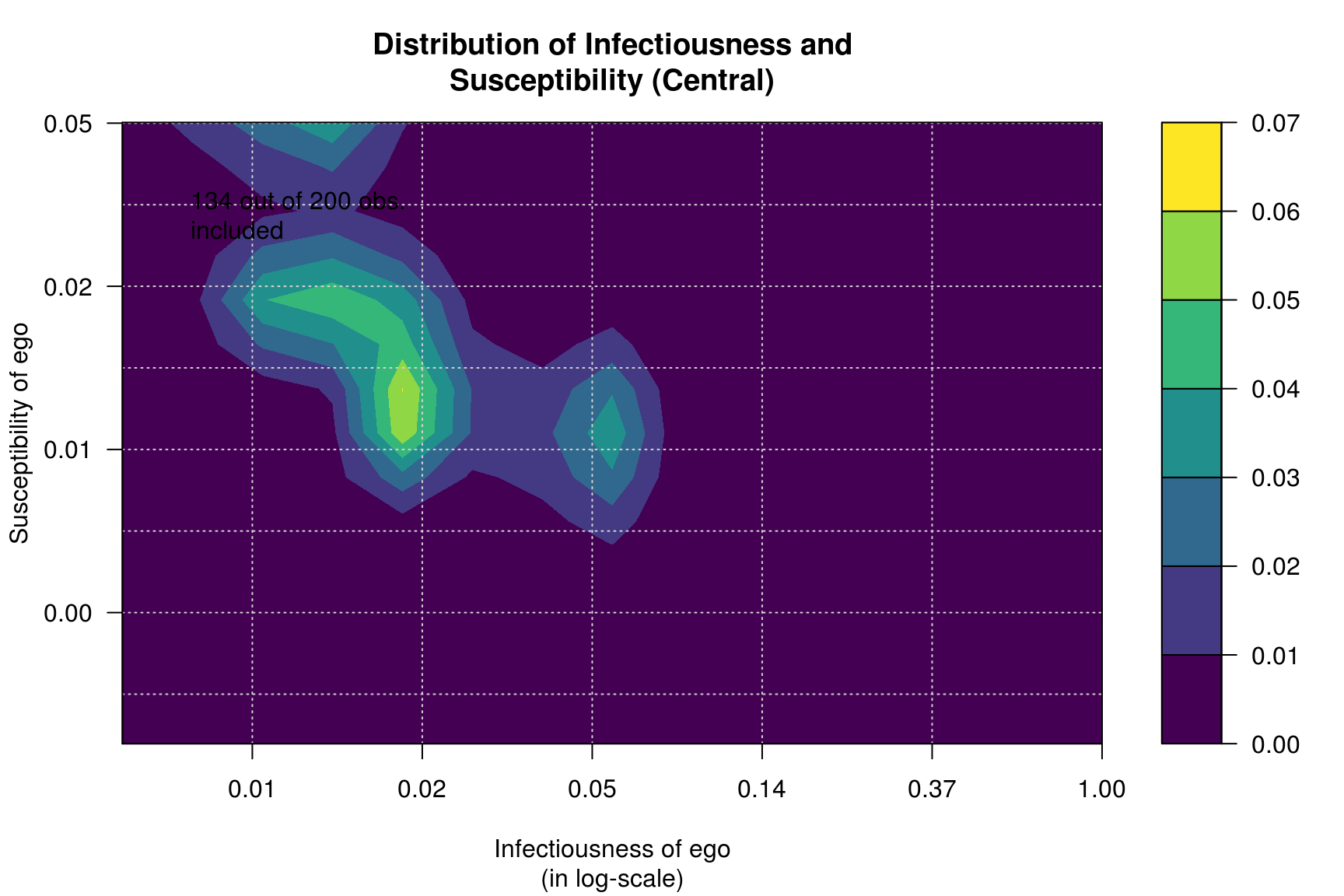
plot_infectsuscep(diffnet_mar, bins=15, K=3,
main = "Distribution of Infectiousness and\nSusceptibility (Marginal)")## Warning in plot_infectsuscep.list(graph$graph, graph$toa, t0, normalize, : When
## applying logscale some observations are missing.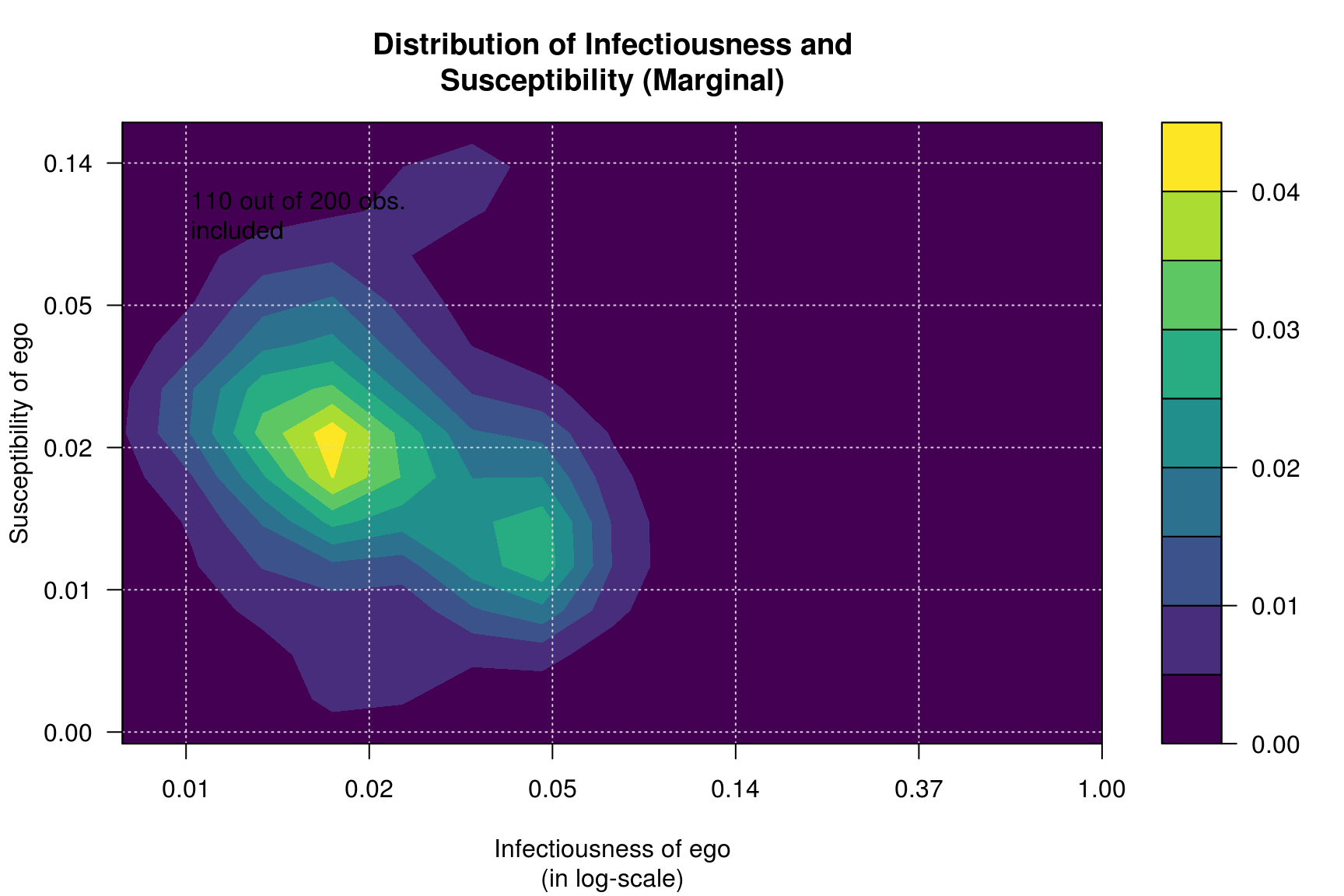
plot_threshold(diffnet_ran)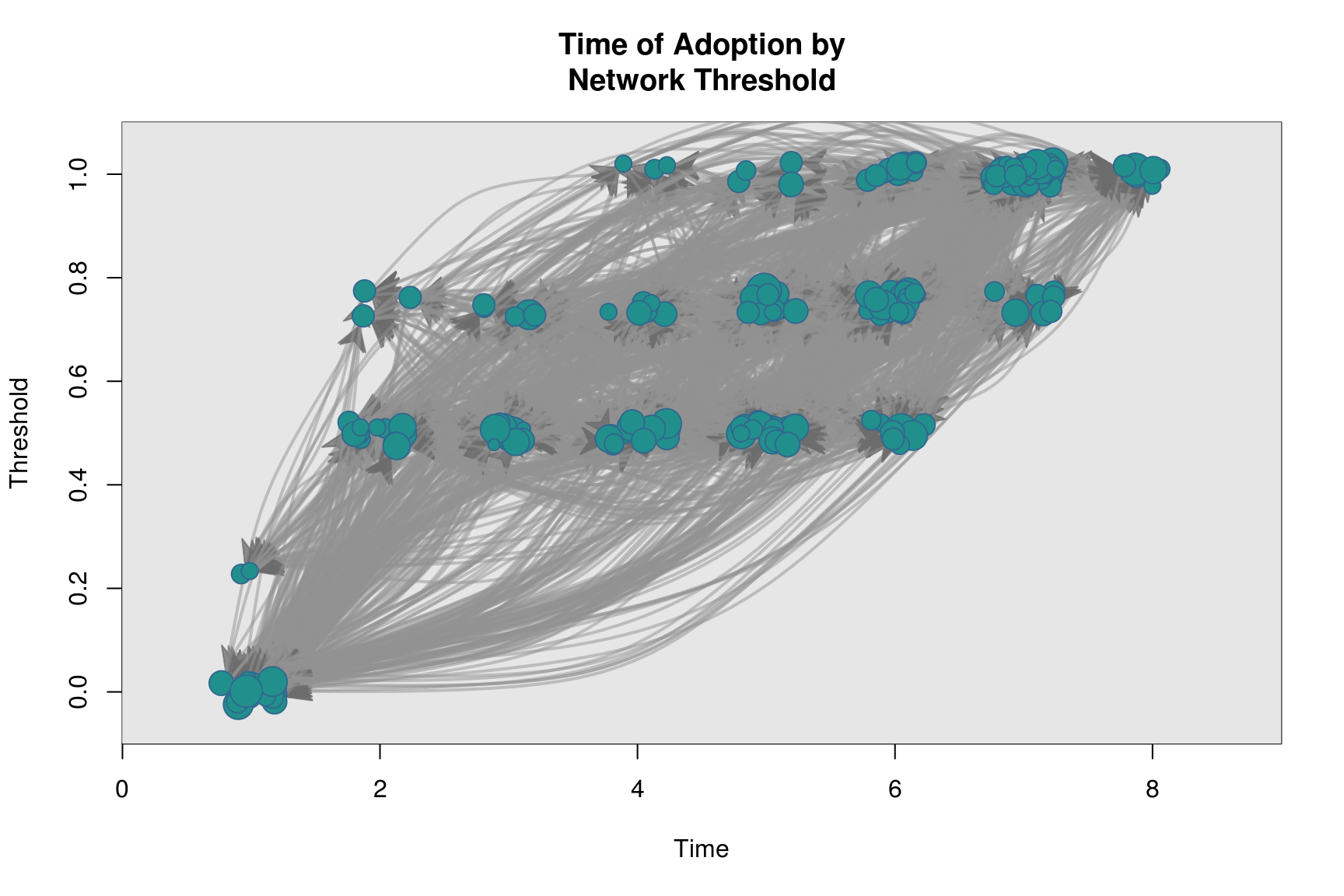
Multiple simulations using rdiffnet_multiple
The rdiffnet_multiple is a wrapper of
rdiffnet that allows performing simulation studies. In
particular, the user can defined a set of shared parameters across
simulations and retrieve one or more statistics from each one of them.
The followin example is included in the manual of the function:
# Simulating a diffusion process with all the defaults but setting
# -seed.nodes- to be random
set.seed(1)
ans0 <- rdiffnet_multiple(R=50, statistic=function(x) sum(!is.na(x$toa)),
n = 100, t = 4, seed.nodes = "random", stop.no.diff=FALSE)## Warning in (function (graph, p, algorithm = "endpoints", both.ends = FALSE, :
## The option -copy.first- is set to TRUE. In this case, the first graph will be
## treated as a baseline, and thus, networks after T=1 will be replaced with T-1.## Warning in (function (graph, p, algorithm = "endpoints", both.ends = FALSE, :
## The option -copy.first- is set to TRUE. In this case, the first graph will be
## treated as a baseline, and thus, networks after T=1 will be replaced with T-1.## Warning in (function (graph, p, algorithm = "endpoints", both.ends = FALSE, :
## The option -copy.first- is set to TRUE. In this case, the first graph will be
## treated as a baseline, and thus, networks after T=1 will be replaced with T-1.## Warning in (function (graph, p, algorithm = "endpoints", both.ends = FALSE, :
## The option -copy.first- is set to TRUE. In this case, the first graph will be
## treated as a baseline, and thus, networks after T=1 will be replaced with T-1.## Warning in (function (graph, p, algorithm = "endpoints", both.ends = FALSE, :
## The option -copy.first- is set to TRUE. In this case, the first graph will be
## treated as a baseline, and thus, networks after T=1 will be replaced with T-1.## Warning in (function (graph, p, algorithm = "endpoints", both.ends = FALSE, :
## The option -copy.first- is set to TRUE. In this case, the first graph will be
## treated as a baseline, and thus, networks after T=1 will be replaced with T-1.## Warning in (function (graph, p, algorithm = "endpoints", both.ends = FALSE, :
## The option -copy.first- is set to TRUE. In this case, the first graph will be
## treated as a baseline, and thus, networks after T=1 will be replaced with T-1.## Warning in (function (graph, p, algorithm = "endpoints", both.ends = FALSE, :
## The option -copy.first- is set to TRUE. In this case, the first graph will be
## treated as a baseline, and thus, networks after T=1 will be replaced with T-1.## Warning in (function (graph, p, algorithm = "endpoints", both.ends = FALSE, :
## The option -copy.first- is set to TRUE. In this case, the first graph will be
## treated as a baseline, and thus, networks after T=1 will be replaced with T-1.## Warning in (function (graph, p, algorithm = "endpoints", both.ends = FALSE, :
## The option -copy.first- is set to TRUE. In this case, the first graph will be
## treated as a baseline, and thus, networks after T=1 will be replaced with T-1.## Warning in (function (graph, p, algorithm = "endpoints", both.ends = FALSE, :
## The option -copy.first- is set to TRUE. In this case, the first graph will be
## treated as a baseline, and thus, networks after T=1 will be replaced with T-1.## Warning in (function (graph, p, algorithm = "endpoints", both.ends = FALSE, :
## The option -copy.first- is set to TRUE. In this case, the first graph will be
## treated as a baseline, and thus, networks after T=1 will be replaced with T-1.## Warning in (function (graph, p, algorithm = "endpoints", both.ends = FALSE, :
## The option -copy.first- is set to TRUE. In this case, the first graph will be
## treated as a baseline, and thus, networks after T=1 will be replaced with T-1.## Warning in (function (graph, p, algorithm = "endpoints", both.ends = FALSE, :
## The option -copy.first- is set to TRUE. In this case, the first graph will be
## treated as a baseline, and thus, networks after T=1 will be replaced with T-1.## Warning in (function (graph, p, algorithm = "endpoints", both.ends = FALSE, :
## The option -copy.first- is set to TRUE. In this case, the first graph will be
## treated as a baseline, and thus, networks after T=1 will be replaced with T-1.## Warning in (function (graph, p, algorithm = "endpoints", both.ends = FALSE, :
## The option -copy.first- is set to TRUE. In this case, the first graph will be
## treated as a baseline, and thus, networks after T=1 will be replaced with T-1.## Warning in (function (graph, p, algorithm = "endpoints", both.ends = FALSE, :
## The option -copy.first- is set to TRUE. In this case, the first graph will be
## treated as a baseline, and thus, networks after T=1 will be replaced with T-1.## Warning in (function (graph, p, algorithm = "endpoints", both.ends = FALSE, :
## The option -copy.first- is set to TRUE. In this case, the first graph will be
## treated as a baseline, and thus, networks after T=1 will be replaced with T-1.## Warning in (function (graph, p, algorithm = "endpoints", both.ends = FALSE, :
## The option -copy.first- is set to TRUE. In this case, the first graph will be
## treated as a baseline, and thus, networks after T=1 will be replaced with T-1.## Warning in (function (graph, p, algorithm = "endpoints", both.ends = FALSE, :
## The option -copy.first- is set to TRUE. In this case, the first graph will be
## treated as a baseline, and thus, networks after T=1 will be replaced with T-1.## Warning in (function (graph, p, algorithm = "endpoints", both.ends = FALSE, :
## The option -copy.first- is set to TRUE. In this case, the first graph will be
## treated as a baseline, and thus, networks after T=1 will be replaced with T-1.## Warning in (function (graph, p, algorithm = "endpoints", both.ends = FALSE, :
## The option -copy.first- is set to TRUE. In this case, the first graph will be
## treated as a baseline, and thus, networks after T=1 will be replaced with T-1.## Warning in (function (graph, p, algorithm = "endpoints", both.ends = FALSE, :
## The option -copy.first- is set to TRUE. In this case, the first graph will be
## treated as a baseline, and thus, networks after T=1 will be replaced with T-1.## Warning in (function (graph, p, algorithm = "endpoints", both.ends = FALSE, :
## The option -copy.first- is set to TRUE. In this case, the first graph will be
## treated as a baseline, and thus, networks after T=1 will be replaced with T-1.## Warning in (function (graph, p, algorithm = "endpoints", both.ends = FALSE, :
## The option -copy.first- is set to TRUE. In this case, the first graph will be
## treated as a baseline, and thus, networks after T=1 will be replaced with T-1.## Warning in (function (graph, p, algorithm = "endpoints", both.ends = FALSE, :
## The option -copy.first- is set to TRUE. In this case, the first graph will be
## treated as a baseline, and thus, networks after T=1 will be replaced with T-1.## Warning in (function (graph, p, algorithm = "endpoints", both.ends = FALSE, :
## The option -copy.first- is set to TRUE. In this case, the first graph will be
## treated as a baseline, and thus, networks after T=1 will be replaced with T-1.## Warning in (function (graph, p, algorithm = "endpoints", both.ends = FALSE, :
## The option -copy.first- is set to TRUE. In this case, the first graph will be
## treated as a baseline, and thus, networks after T=1 will be replaced with T-1.## Warning in (function (graph, p, algorithm = "endpoints", both.ends = FALSE, :
## The option -copy.first- is set to TRUE. In this case, the first graph will be
## treated as a baseline, and thus, networks after T=1 will be replaced with T-1.## Warning in (function (graph, p, algorithm = "endpoints", both.ends = FALSE, :
## The option -copy.first- is set to TRUE. In this case, the first graph will be
## treated as a baseline, and thus, networks after T=1 will be replaced with T-1.## Warning in (function (graph, p, algorithm = "endpoints", both.ends = FALSE, :
## The option -copy.first- is set to TRUE. In this case, the first graph will be
## treated as a baseline, and thus, networks after T=1 will be replaced with T-1.## Warning in (function (graph, p, algorithm = "endpoints", both.ends = FALSE, :
## The option -copy.first- is set to TRUE. In this case, the first graph will be
## treated as a baseline, and thus, networks after T=1 will be replaced with T-1.## Warning in (function (graph, p, algorithm = "endpoints", both.ends = FALSE, :
## The option -copy.first- is set to TRUE. In this case, the first graph will be
## treated as a baseline, and thus, networks after T=1 will be replaced with T-1.## Warning in (function (graph, p, algorithm = "endpoints", both.ends = FALSE, :
## The option -copy.first- is set to TRUE. In this case, the first graph will be
## treated as a baseline, and thus, networks after T=1 will be replaced with T-1.## Warning in (function (graph, p, algorithm = "endpoints", both.ends = FALSE, :
## The option -copy.first- is set to TRUE. In this case, the first graph will be
## treated as a baseline, and thus, networks after T=1 will be replaced with T-1.## Warning in (function (graph, p, algorithm = "endpoints", both.ends = FALSE, :
## The option -copy.first- is set to TRUE. In this case, the first graph will be
## treated as a baseline, and thus, networks after T=1 will be replaced with T-1.## Warning in (function (graph, p, algorithm = "endpoints", both.ends = FALSE, :
## The option -copy.first- is set to TRUE. In this case, the first graph will be
## treated as a baseline, and thus, networks after T=1 will be replaced with T-1.## Warning in (function (graph, p, algorithm = "endpoints", both.ends = FALSE, :
## The option -copy.first- is set to TRUE. In this case, the first graph will be
## treated as a baseline, and thus, networks after T=1 will be replaced with T-1.## Warning in (function (graph, p, algorithm = "endpoints", both.ends = FALSE, :
## The option -copy.first- is set to TRUE. In this case, the first graph will be
## treated as a baseline, and thus, networks after T=1 will be replaced with T-1.## Warning in (function (graph, p, algorithm = "endpoints", both.ends = FALSE, :
## The option -copy.first- is set to TRUE. In this case, the first graph will be
## treated as a baseline, and thus, networks after T=1 will be replaced with T-1.## Warning in (function (graph, p, algorithm = "endpoints", both.ends = FALSE, :
## The option -copy.first- is set to TRUE. In this case, the first graph will be
## treated as a baseline, and thus, networks after T=1 will be replaced with T-1.## Warning in (function (graph, p, algorithm = "endpoints", both.ends = FALSE, :
## The option -copy.first- is set to TRUE. In this case, the first graph will be
## treated as a baseline, and thus, networks after T=1 will be replaced with T-1.## Warning in (function (graph, p, algorithm = "endpoints", both.ends = FALSE, :
## The option -copy.first- is set to TRUE. In this case, the first graph will be
## treated as a baseline, and thus, networks after T=1 will be replaced with T-1.## Warning in (function (graph, p, algorithm = "endpoints", both.ends = FALSE, :
## The option -copy.first- is set to TRUE. In this case, the first graph will be
## treated as a baseline, and thus, networks after T=1 will be replaced with T-1.## Warning in (function (graph, p, algorithm = "endpoints", both.ends = FALSE, :
## The option -copy.first- is set to TRUE. In this case, the first graph will be
## treated as a baseline, and thus, networks after T=1 will be replaced with T-1.## Warning in (function (graph, p, algorithm = "endpoints", both.ends = FALSE, :
## The option -copy.first- is set to TRUE. In this case, the first graph will be
## treated as a baseline, and thus, networks after T=1 will be replaced with T-1.## Warning in (function (graph, p, algorithm = "endpoints", both.ends = FALSE, :
## The option -copy.first- is set to TRUE. In this case, the first graph will be
## treated as a baseline, and thus, networks after T=1 will be replaced with T-1.## Warning in (function (graph, p, algorithm = "endpoints", both.ends = FALSE, :
## The option -copy.first- is set to TRUE. In this case, the first graph will be
## treated as a baseline, and thus, networks after T=1 will be replaced with T-1.## Warning in (function (graph, p, algorithm = "endpoints", both.ends = FALSE, :
## The option -copy.first- is set to TRUE. In this case, the first graph will be
## treated as a baseline, and thus, networks after T=1 will be replaced with T-1.## Warning in (function (graph, p, algorithm = "endpoints", both.ends = FALSE, :
## The option -copy.first- is set to TRUE. In this case, the first graph will be
## treated as a baseline, and thus, networks after T=1 will be replaced with T-1.## Warning in netdiffuseR::rdiffnet(...): No diffusion in this network.
# Simulating a diffusion process with all the defaults but setting
# -seed.nodes- to be central
set.seed(1)
ans1 <- rdiffnet_multiple(R=50, statistic=function(x) sum(!is.na(x$toa)),
n = 100, t = 4, seed.nodes = "central", stop.no.diff=FALSE)## Warning in (function (graph, p, algorithm = "endpoints", both.ends = FALSE, :
## The option -copy.first- is set to TRUE. In this case, the first graph will be
## treated as a baseline, and thus, networks after T=1 will be replaced with T-1.## Warning in (function (graph, p, algorithm = "endpoints", both.ends = FALSE, :
## The option -copy.first- is set to TRUE. In this case, the first graph will be
## treated as a baseline, and thus, networks after T=1 will be replaced with T-1.## Warning in (function (graph, p, algorithm = "endpoints", both.ends = FALSE, :
## The option -copy.first- is set to TRUE. In this case, the first graph will be
## treated as a baseline, and thus, networks after T=1 will be replaced with T-1.## Warning in (function (graph, p, algorithm = "endpoints", both.ends = FALSE, :
## The option -copy.first- is set to TRUE. In this case, the first graph will be
## treated as a baseline, and thus, networks after T=1 will be replaced with T-1.## Warning in (function (graph, p, algorithm = "endpoints", both.ends = FALSE, :
## The option -copy.first- is set to TRUE. In this case, the first graph will be
## treated as a baseline, and thus, networks after T=1 will be replaced with T-1.## Warning in (function (graph, p, algorithm = "endpoints", both.ends = FALSE, :
## The option -copy.first- is set to TRUE. In this case, the first graph will be
## treated as a baseline, and thus, networks after T=1 will be replaced with T-1.## Warning in (function (graph, p, algorithm = "endpoints", both.ends = FALSE, :
## The option -copy.first- is set to TRUE. In this case, the first graph will be
## treated as a baseline, and thus, networks after T=1 will be replaced with T-1.## Warning in (function (graph, p, algorithm = "endpoints", both.ends = FALSE, :
## The option -copy.first- is set to TRUE. In this case, the first graph will be
## treated as a baseline, and thus, networks after T=1 will be replaced with T-1.## Warning in (function (graph, p, algorithm = "endpoints", both.ends = FALSE, :
## The option -copy.first- is set to TRUE. In this case, the first graph will be
## treated as a baseline, and thus, networks after T=1 will be replaced with T-1.## Warning in (function (graph, p, algorithm = "endpoints", both.ends = FALSE, :
## The option -copy.first- is set to TRUE. In this case, the first graph will be
## treated as a baseline, and thus, networks after T=1 will be replaced with T-1.## Warning in (function (graph, p, algorithm = "endpoints", both.ends = FALSE, :
## The option -copy.first- is set to TRUE. In this case, the first graph will be
## treated as a baseline, and thus, networks after T=1 will be replaced with T-1.## Warning in (function (graph, p, algorithm = "endpoints", both.ends = FALSE, :
## The option -copy.first- is set to TRUE. In this case, the first graph will be
## treated as a baseline, and thus, networks after T=1 will be replaced with T-1.## Warning in (function (graph, p, algorithm = "endpoints", both.ends = FALSE, :
## The option -copy.first- is set to TRUE. In this case, the first graph will be
## treated as a baseline, and thus, networks after T=1 will be replaced with T-1.## Warning in (function (graph, p, algorithm = "endpoints", both.ends = FALSE, :
## The option -copy.first- is set to TRUE. In this case, the first graph will be
## treated as a baseline, and thus, networks after T=1 will be replaced with T-1.## Warning in (function (graph, p, algorithm = "endpoints", both.ends = FALSE, :
## The option -copy.first- is set to TRUE. In this case, the first graph will be
## treated as a baseline, and thus, networks after T=1 will be replaced with T-1.## Warning in (function (graph, p, algorithm = "endpoints", both.ends = FALSE, :
## The option -copy.first- is set to TRUE. In this case, the first graph will be
## treated as a baseline, and thus, networks after T=1 will be replaced with T-1.## Warning in (function (graph, p, algorithm = "endpoints", both.ends = FALSE, :
## The option -copy.first- is set to TRUE. In this case, the first graph will be
## treated as a baseline, and thus, networks after T=1 will be replaced with T-1.## Warning in (function (graph, p, algorithm = "endpoints", both.ends = FALSE, :
## The option -copy.first- is set to TRUE. In this case, the first graph will be
## treated as a baseline, and thus, networks after T=1 will be replaced with T-1.## Warning in (function (graph, p, algorithm = "endpoints", both.ends = FALSE, :
## The option -copy.first- is set to TRUE. In this case, the first graph will be
## treated as a baseline, and thus, networks after T=1 will be replaced with T-1.## Warning in (function (graph, p, algorithm = "endpoints", both.ends = FALSE, :
## The option -copy.first- is set to TRUE. In this case, the first graph will be
## treated as a baseline, and thus, networks after T=1 will be replaced with T-1.## Warning in (function (graph, p, algorithm = "endpoints", both.ends = FALSE, :
## The option -copy.first- is set to TRUE. In this case, the first graph will be
## treated as a baseline, and thus, networks after T=1 will be replaced with T-1.## Warning in (function (graph, p, algorithm = "endpoints", both.ends = FALSE, :
## The option -copy.first- is set to TRUE. In this case, the first graph will be
## treated as a baseline, and thus, networks after T=1 will be replaced with T-1.## Warning in (function (graph, p, algorithm = "endpoints", both.ends = FALSE, :
## The option -copy.first- is set to TRUE. In this case, the first graph will be
## treated as a baseline, and thus, networks after T=1 will be replaced with T-1.## Warning in (function (graph, p, algorithm = "endpoints", both.ends = FALSE, :
## The option -copy.first- is set to TRUE. In this case, the first graph will be
## treated as a baseline, and thus, networks after T=1 will be replaced with T-1.## Warning in (function (graph, p, algorithm = "endpoints", both.ends = FALSE, :
## The option -copy.first- is set to TRUE. In this case, the first graph will be
## treated as a baseline, and thus, networks after T=1 will be replaced with T-1.## Warning in (function (graph, p, algorithm = "endpoints", both.ends = FALSE, :
## The option -copy.first- is set to TRUE. In this case, the first graph will be
## treated as a baseline, and thus, networks after T=1 will be replaced with T-1.## Warning in (function (graph, p, algorithm = "endpoints", both.ends = FALSE, :
## The option -copy.first- is set to TRUE. In this case, the first graph will be
## treated as a baseline, and thus, networks after T=1 will be replaced with T-1.## Warning in (function (graph, p, algorithm = "endpoints", both.ends = FALSE, :
## The option -copy.first- is set to TRUE. In this case, the first graph will be
## treated as a baseline, and thus, networks after T=1 will be replaced with T-1.## Warning in (function (graph, p, algorithm = "endpoints", both.ends = FALSE, :
## The option -copy.first- is set to TRUE. In this case, the first graph will be
## treated as a baseline, and thus, networks after T=1 will be replaced with T-1.## Warning in (function (graph, p, algorithm = "endpoints", both.ends = FALSE, :
## The option -copy.first- is set to TRUE. In this case, the first graph will be
## treated as a baseline, and thus, networks after T=1 will be replaced with T-1.## Warning in (function (graph, p, algorithm = "endpoints", both.ends = FALSE, :
## The option -copy.first- is set to TRUE. In this case, the first graph will be
## treated as a baseline, and thus, networks after T=1 will be replaced with T-1.## Warning in (function (graph, p, algorithm = "endpoints", both.ends = FALSE, :
## The option -copy.first- is set to TRUE. In this case, the first graph will be
## treated as a baseline, and thus, networks after T=1 will be replaced with T-1.## Warning in (function (graph, p, algorithm = "endpoints", both.ends = FALSE, :
## The option -copy.first- is set to TRUE. In this case, the first graph will be
## treated as a baseline, and thus, networks after T=1 will be replaced with T-1.## Warning in (function (graph, p, algorithm = "endpoints", both.ends = FALSE, :
## The option -copy.first- is set to TRUE. In this case, the first graph will be
## treated as a baseline, and thus, networks after T=1 will be replaced with T-1.## Warning in (function (graph, p, algorithm = "endpoints", both.ends = FALSE, :
## The option -copy.first- is set to TRUE. In this case, the first graph will be
## treated as a baseline, and thus, networks after T=1 will be replaced with T-1.## Warning in (function (graph, p, algorithm = "endpoints", both.ends = FALSE, :
## The option -copy.first- is set to TRUE. In this case, the first graph will be
## treated as a baseline, and thus, networks after T=1 will be replaced with T-1.## Warning in (function (graph, p, algorithm = "endpoints", both.ends = FALSE, :
## The option -copy.first- is set to TRUE. In this case, the first graph will be
## treated as a baseline, and thus, networks after T=1 will be replaced with T-1.## Warning in (function (graph, p, algorithm = "endpoints", both.ends = FALSE, :
## The option -copy.first- is set to TRUE. In this case, the first graph will be
## treated as a baseline, and thus, networks after T=1 will be replaced with T-1.## Warning in (function (graph, p, algorithm = "endpoints", both.ends = FALSE, :
## The option -copy.first- is set to TRUE. In this case, the first graph will be
## treated as a baseline, and thus, networks after T=1 will be replaced with T-1.## Warning in (function (graph, p, algorithm = "endpoints", both.ends = FALSE, :
## The option -copy.first- is set to TRUE. In this case, the first graph will be
## treated as a baseline, and thus, networks after T=1 will be replaced with T-1.## Warning in (function (graph, p, algorithm = "endpoints", both.ends = FALSE, :
## The option -copy.first- is set to TRUE. In this case, the first graph will be
## treated as a baseline, and thus, networks after T=1 will be replaced with T-1.## Warning in (function (graph, p, algorithm = "endpoints", both.ends = FALSE, :
## The option -copy.first- is set to TRUE. In this case, the first graph will be
## treated as a baseline, and thus, networks after T=1 will be replaced with T-1.## Warning in (function (graph, p, algorithm = "endpoints", both.ends = FALSE, :
## The option -copy.first- is set to TRUE. In this case, the first graph will be
## treated as a baseline, and thus, networks after T=1 will be replaced with T-1.## Warning in (function (graph, p, algorithm = "endpoints", both.ends = FALSE, :
## The option -copy.first- is set to TRUE. In this case, the first graph will be
## treated as a baseline, and thus, networks after T=1 will be replaced with T-1.## Warning in (function (graph, p, algorithm = "endpoints", both.ends = FALSE, :
## The option -copy.first- is set to TRUE. In this case, the first graph will be
## treated as a baseline, and thus, networks after T=1 will be replaced with T-1.## Warning in (function (graph, p, algorithm = "endpoints", both.ends = FALSE, :
## The option -copy.first- is set to TRUE. In this case, the first graph will be
## treated as a baseline, and thus, networks after T=1 will be replaced with T-1.## Warning in (function (graph, p, algorithm = "endpoints", both.ends = FALSE, :
## The option -copy.first- is set to TRUE. In this case, the first graph will be
## treated as a baseline, and thus, networks after T=1 will be replaced with T-1.## Warning in (function (graph, p, algorithm = "endpoints", both.ends = FALSE, :
## The option -copy.first- is set to TRUE. In this case, the first graph will be
## treated as a baseline, and thus, networks after T=1 will be replaced with T-1.## Warning in (function (graph, p, algorithm = "endpoints", both.ends = FALSE, :
## The option -copy.first- is set to TRUE. In this case, the first graph will be
## treated as a baseline, and thus, networks after T=1 will be replaced with T-1.## Warning in (function (graph, p, algorithm = "endpoints", both.ends = FALSE, :
## The option -copy.first- is set to TRUE. In this case, the first graph will be
## treated as a baseline, and thus, networks after T=1 will be replaced with T-1.
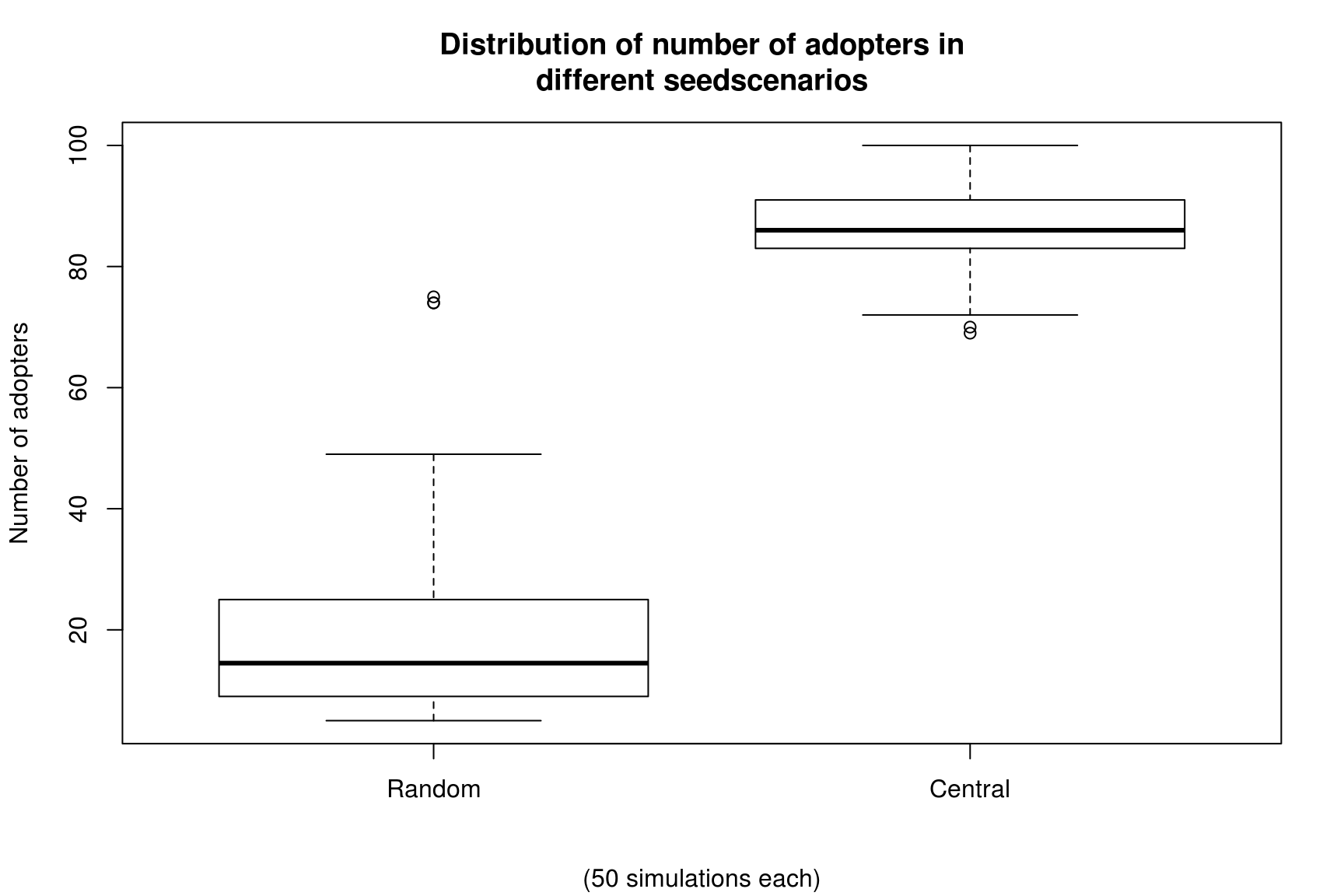
boxplot(cbind(Random = ans0, Central = ans1),
main="Distribution of number of adopters in\ndifferent seedscenarios",
sub = "(50 simulations each)", ylab="Number of adopters")