Generates adjacency matrix from an edgelist and vice versa.
edgelist_to_adjmat(
edgelist,
w = NULL,
t0 = NULL,
t1 = NULL,
t = NULL,
simplify = TRUE,
undirected = getOption("diffnet.undirected"),
self = getOption("diffnet.self"),
multiple = getOption("diffnet.multiple"),
keep.isolates = TRUE,
recode.ids = TRUE
)
adjmat_to_edgelist(
graph,
undirected = getOption("diffnet.undirected", FALSE),
keep.isolates = getOption("diffnet.keep.isolates", TRUE)
)Arguments
- edgelist
Two column matrix/data.frame in the form of ego -source- and alter -target- (see details).
- w
Numeric vector. Strength of ties (optional).
- t0
Integer vector. Starting time of the ties (optional).
- t1
Integer vector. Finishing time of the ties (optional).
- t
Integer scalar. Repeat the network
ttimes (if not0,t1are provided).- simplify
Logical scalar. When TRUE and
times=NULLit will return an adjacency matrix, otherwise an array of adjacency matrices. (see details).- undirected
Logical scalar. When
TRUEonly the lower triangle of the adjacency matrix will considered (faster).- self
Logical scalar. When
TRUEautolinks (loops, self edges) are allowed (see details).- multiple
Logical scalar. When
TRUEallows multiple edges.- keep.isolates
Logical scalar. When FALSE, rows with
NA/NULLvalues (isolated vertices unless have autolink) will be droped (see details).- recode.ids
Logical scalar. When TRUE ids are recoded using
as.factor(see details).- graph
Any class of accepted graph format (see
netdiffuseR-graphs).
Value
In the case of edgelist_to_adjmat either an adjacency matrix
(if times is NULL) or an array of these (if times is not null). For
adjmat_to_edgelist the output is an edgelist with the following columns:
- ego
Origin of the tie.
- alter
Target of the tie.
- value
Value in the adjacency matrix.
- time
Either a 1 (if the network is static) or the time stamp of the tie.
Details
When converting from edglist to adjmat the function will recode the
edgelist before starting. The user can keep track after the recording by checking
the resulting adjacency matrices' row.names. In the case that the
user decides skipping the recoding (because wants to keep vertices index numbers,
implying that the resulting graph will have isolated vertices), he can override
this by setting recode.ids=FALSE (see example).
When multiple edges are included, multiple=TRUE,each vertex between \(\{i,j\}\) will be counted
as many times it appears in the edgelist. So if a vertex \(\{i,j\}\) appears 2
times, the adjacency matrix element (i,j) will be 2.
Edges with incomplete information (missing data on w or times) are
not included on the graph. Incomplete cases are tagged using complete.cases
and can be retrieved by the user by accessing the attribute incomplete.
Were the case that either ego or alter are missing (i.e. NA values), the
function will either way include the non-missing vertex. See below for an example
of this.
The function performs several checks before starting to create the adjacency matrix. These are:
Dimensions of the inputs, such as number of columns and length of vectors
Having complete cases. If anly edge has a non-numeric value such as NAs or NULL in either
timesorw, it will be removed. A full list of such edges can be retrieved from the attributeincompleteNodes and times ids coding
recode.ids=FALSE is useful when the vertices ids have already been
coded. For example, after having use adjmat_to_edgelist, ids are
correctly encoded, so when going back (using edgelist_to_adjmat)
recode.ids should be FALSE.
See also
Other data management functions:
diffnet-class,
egonet_attrs(),
isolated(),
survey_to_diffnet()
Examples
# Base data
set.seed(123)
n <- 5
edgelist <- rgraph_er(n, as.edgelist=TRUE, p=.2)[,c("ego","alter")]
times <- sample.int(3, nrow(edgelist), replace=TRUE)
w <- abs(rnorm(nrow(edgelist)))
# Simple example
edgelist_to_adjmat(edgelist)
#> 5 x 5 sparse Matrix of class "dgCMatrix"
#> 1 2 3 4 5
#> 1 . . . . 1
#> 2 1 . . . 1
#> 3 . . . 1 .
#> 4 . . . . 1
#> 5 . . . 1 .
edgelist_to_adjmat(edgelist, undirected = TRUE)
#> 5 x 5 sparse Matrix of class "dgCMatrix"
#> 1 2 3 4 5
#> 1 . 1 . . 1
#> 2 1 . . . 1
#> 3 . . . 1 .
#> 4 . . 1 . 1
#> 5 1 1 . 1 .
# Using w
edgelist_to_adjmat(edgelist, w)
#> 5 x 5 sparse Matrix of class "dgCMatrix"
#> 1 2 3 4 5
#> 1 . . . . 1.9666172
#> 2 0.5558411 . . . 0.7013559
#> 3 . . . 1.7869131 .
#> 4 . . . . 0.4727914
#> 5 . . . 0.4978505 .
edgelist_to_adjmat(edgelist, w, undirected = TRUE)
#> 5 x 5 sparse Matrix of class "dgCMatrix"
#> 1 2 3 4 5
#> 1 . 0.5558411 . . 1.9666172
#> 2 0.5558411 . . . 0.7013559
#> 3 . . . 1.7869131 .
#> 4 . . 1.786913 . 0.4978505
#> 5 1.9666172 0.7013559 . 0.4978505 .
# Using times
edgelist_to_adjmat(edgelist, t0 = times)
#> $`1`
#> 5 x 5 sparse Matrix of class "dgCMatrix"
#> 1 2 3 4 5
#> 1 . . . . 1
#> 2 1 . . . .
#> 3 . . . 1 .
#> 4 . . . . .
#> 5 . . . 1 .
#>
#> $`2`
#> 5 x 5 sparse Matrix of class "dgCMatrix"
#> 1 2 3 4 5
#> 1 . . . . 1
#> 2 1 . . . .
#> 3 . . . 1 .
#> 4 . . . . 1
#> 5 . . . 1 .
#>
#> $`3`
#> 5 x 5 sparse Matrix of class "dgCMatrix"
#> 1 2 3 4 5
#> 1 . . . . 1
#> 2 1 . . . 1
#> 3 . . . 1 .
#> 4 . . . . 1
#> 5 . . . 1 .
#>
#> attr(,"incomplete")
#> integer(0)
edgelist_to_adjmat(edgelist, t0 = times, undirected = TRUE)
#> $`1`
#> 5 x 5 sparse Matrix of class "dgCMatrix"
#> 1 2 3 4 5
#> 1 . 1 . . 1
#> 2 1 . . . .
#> 3 . . . 1 .
#> 4 . . 1 . 1
#> 5 1 . . 1 .
#>
#> $`2`
#> 5 x 5 sparse Matrix of class "dgCMatrix"
#> 1 2 3 4 5
#> 1 . 1 . . 1
#> 2 1 . . . .
#> 3 . . . 1 .
#> 4 . . 1 . 1
#> 5 1 . . 1 .
#>
#> $`3`
#> 5 x 5 sparse Matrix of class "dgCMatrix"
#> 1 2 3 4 5
#> 1 . 1 . . 1
#> 2 1 . . . 1
#> 3 . . . 1 .
#> 4 . . 1 . 1
#> 5 1 1 . 1 .
#>
#> attr(,"incomplete")
#> integer(0)
# Using times and w
edgelist_to_adjmat(edgelist, t0 = times, w = w)
#> $`1`
#> 5 x 5 sparse Matrix of class "dgCMatrix"
#> 1 2 3 4 5
#> 1 . . . . 1.966617
#> 2 0.5558411 . . . .
#> 3 . . . 1.7869131 .
#> 4 . . . . .
#> 5 . . . 0.4978505 .
#>
#> $`2`
#> 5 x 5 sparse Matrix of class "dgCMatrix"
#> 1 2 3 4 5
#> 1 . . . . 1.9666172
#> 2 0.5558411 . . . .
#> 3 . . . 1.7869131 .
#> 4 . . . . 0.4727914
#> 5 . . . 0.4978505 .
#>
#> $`3`
#> 5 x 5 sparse Matrix of class "dgCMatrix"
#> 1 2 3 4 5
#> 1 . . . . 1.9666172
#> 2 0.5558411 . . . 0.7013559
#> 3 . . . 1.7869131 .
#> 4 . . . . 0.4727914
#> 5 . . . 0.4978505 .
#>
#> attr(,"incomplete")
#> integer(0)
edgelist_to_adjmat(edgelist, t0 = times, undirected = TRUE, w = w)
#> $`1`
#> 5 x 5 sparse Matrix of class "dgCMatrix"
#> 1 2 3 4 5
#> 1 . 0.5558411 . . 1.9666172
#> 2 0.5558411 . . . .
#> 3 . . . 1.7869131 .
#> 4 . . 1.786913 . 0.4978505
#> 5 1.9666172 . . 0.4978505 .
#>
#> $`2`
#> 5 x 5 sparse Matrix of class "dgCMatrix"
#> 1 2 3 4 5
#> 1 . 0.5558411 . . 1.9666172
#> 2 0.5558411 . . . .
#> 3 . . . 1.7869131 .
#> 4 . . 1.786913 . 0.4978505
#> 5 1.9666172 . . 0.4978505 .
#>
#> $`3`
#> 5 x 5 sparse Matrix of class "dgCMatrix"
#> 1 2 3 4 5
#> 1 . 0.5558411 . . 1.9666172
#> 2 0.5558411 . . . 0.7013559
#> 3 . . . 1.7869131 .
#> 4 . . 1.786913 . 0.4978505
#> 5 1.9666172 0.7013559 . 0.4978505 .
#>
#> attr(,"incomplete")
#> integer(0)
# Not recoding ----------------------------------------------------
# Notice that vertices 3, 4 and 5 are not present in this graph.
graph <- matrix(c(
1,2,6,
6,6,7
), ncol=2)
# Generates an adjmat of size 4 x 4
edgelist_to_adjmat(graph)
#> 4 x 4 sparse Matrix of class "dgCMatrix"
#> 1 2 6 7
#> 1 . . 1 .
#> 2 . . 1 .
#> 6 . . . 1
#> 7 . . . .
# Generates an adjmat of size 7 x 7
edgelist_to_adjmat(graph, recode.ids=FALSE)
#> 7 x 7 sparse Matrix of class "dgCMatrix"
#> 1 2 3 4 5 6 7
#> 1 . . . . . 1 .
#> 2 . . . . . 1 .
#> 3 . . . . . . .
#> 4 . . . . . . .
#> 5 . . . . . . .
#> 6 . . . . . . 1
#> 7 . . . . . . .
# Dynamic with spells -------------------------------------------------------
edgelist <- rbind(
c(1,2,NA,1990),
c(2,3,NA,1991),
c(3,4,1991,1992),
c(4,1,1992,1993),
c(1,2,1993,1993)
)
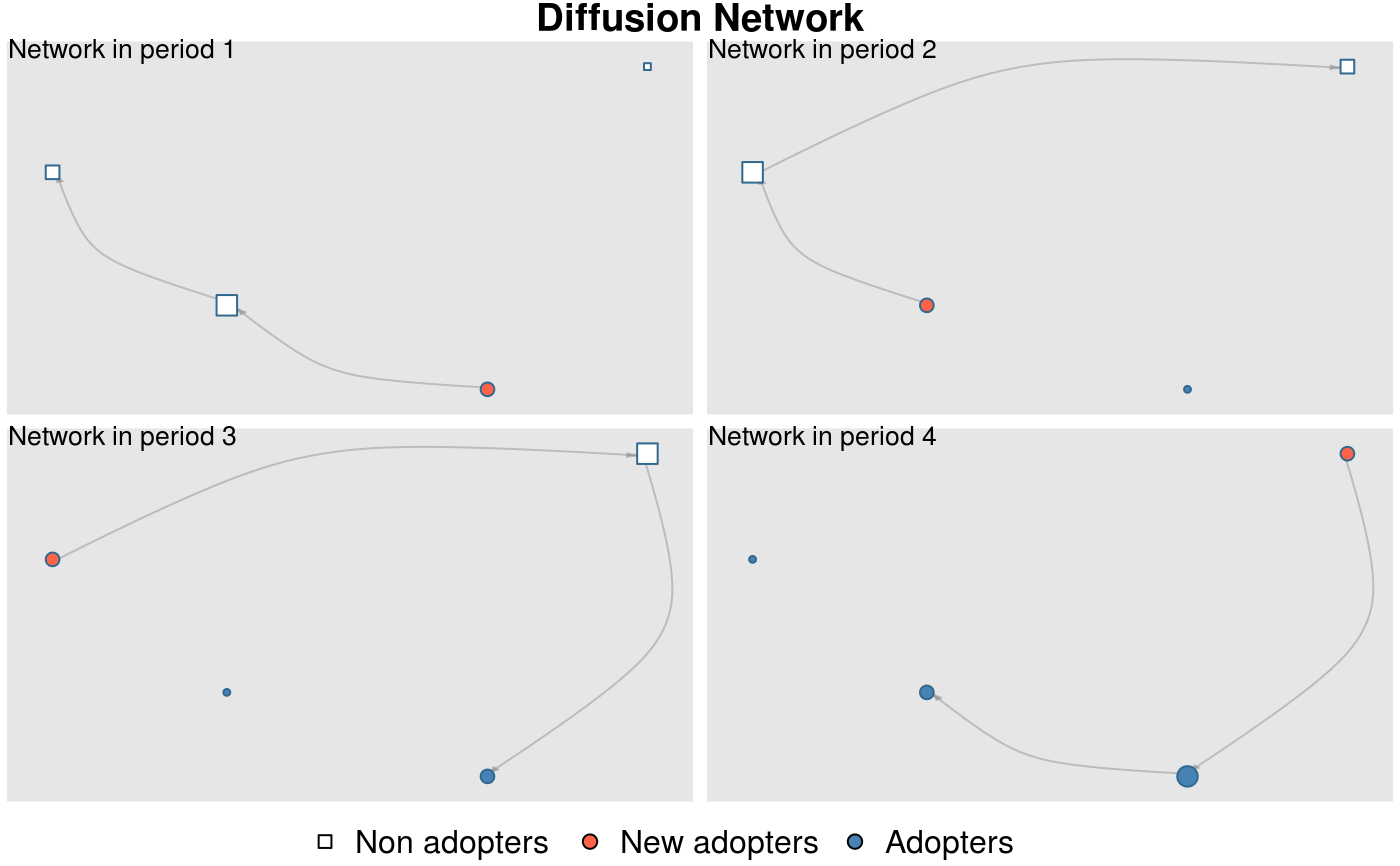
graph <- edgelist_to_adjmat(edgelist[,1:2], t0=edgelist[,3], t1=edgelist[,4])
# Creating a diffnet object with it so we can apply the plot_diffnet function
diffnet <- as_diffnet(graph, toa=1:4)
plot_diffnet(diffnet, label=rownames(diffnet))
 # Missing alter in the edgelist ---------------------------------------------
data(fakeEdgelist)
# Notice that edge 202 is isolated
fakeEdgelist
#> ego alter value
#> 1 102 101 1
#> 2 103 102 1
#> 3 102 103 1
#> 4 105 103 1
#> 5 105 104 2
#> 6 104 105 1
#> 7 205 201 1
#> 8 210 201 1
#> 9 210 205 1
#> 10 205 210 1
#> 11 202 <NA> NA
# The function still includes vertex 202
edgelist_to_adjmat(fakeEdgelist[,1:2])
#> 9 x 9 sparse Matrix of class "dgCMatrix"
#> 101 102 103 104 105 201 202 205 210
#> 101 . . . . . . . . .
#> 102 1 . 1 . . . . . .
#> 103 . 1 . . . . . . .
#> 104 . . . . 1 . . . .
#> 105 . . 1 1 . . . . .
#> 201 . . . . . . . . .
#> 202 . . . . . . . . .
#> 205 . . . . . 1 . . 1
#> 210 . . . . . 1 . 1 .
edgelist
#> [,1] [,2] [,3] [,4]
#> [1,] 1 2 NA 1990
#> [2,] 2 3 NA 1991
#> [3,] 3 4 1991 1992
#> [4,] 4 1 1992 1993
#> [5,] 1 2 1993 1993
# Missing alter in the edgelist ---------------------------------------------
data(fakeEdgelist)
# Notice that edge 202 is isolated
fakeEdgelist
#> ego alter value
#> 1 102 101 1
#> 2 103 102 1
#> 3 102 103 1
#> 4 105 103 1
#> 5 105 104 2
#> 6 104 105 1
#> 7 205 201 1
#> 8 210 201 1
#> 9 210 205 1
#> 10 205 210 1
#> 11 202 <NA> NA
# The function still includes vertex 202
edgelist_to_adjmat(fakeEdgelist[,1:2])
#> 9 x 9 sparse Matrix of class "dgCMatrix"
#> 101 102 103 104 105 201 202 205 210
#> 101 . . . . . . . . .
#> 102 1 . 1 . . . . . .
#> 103 . 1 . . . . . . .
#> 104 . . . . 1 . . . .
#> 105 . . 1 1 . . . . .
#> 201 . . . . . . . . .
#> 202 . . . . . . . . .
#> 205 . . . . . 1 . . 1
#> 210 . . . . . 1 . 1 .
edgelist
#> [,1] [,2] [,3] [,4]
#> [1,] 1 2 NA 1990
#> [2,] 2 3 NA 1991
#> [3,] 3 4 1991 1992
#> [4,] 4 1 1992 1993
#> [5,] 1 2 1993 1993