Convert survey-like data and edgelists to a diffnet object
Source: R/survey_to_diffnet.R
survey_to_diffnet.RdThese convenient functions turn network nomination datasets and edgelists with
vertex attributes datasets into diffnet objects. Both work as wrappers of
edgelist_to_adjmat and new_diffnet.
survey_to_diffnet(
dat,
idvar,
netvars,
toavar,
groupvar = NULL,
no.unsurveyed = TRUE,
timevar = NULL,
t = NULL,
undirected = getOption("diffnet.undirected", FALSE),
self = getOption("diffnet.self", FALSE),
multiple = getOption("diffnet.multiple", FALSE),
keep.isolates = TRUE,
recode.ids = TRUE,
warn.coercion = TRUE,
...
)
edgelist_to_diffnet(
edgelist,
w = NULL,
t0 = NULL,
t1 = NULL,
dat,
idvar,
toavar,
timevar = NULL,
undirected = getOption("diffnet.undirected", FALSE),
self = getOption("diffnet.self", FALSE),
multiple = getOption("diffnet.multiple", FALSE),
fill.missing = NULL,
keep.isolates = TRUE,
recode.ids = TRUE,
warn.coercion = TRUE
)Arguments
- dat
A data frame.
- idvar
Character scalar. Name of the id variable.
- netvars
Character vector. Names of the network nomination variables.
- toavar
Character scalar. Name of the time of adoption variable.
- groupvar
Character scalar. Name of cohort variable (e.g. city).
- no.unsurveyed
Logical scalar. When
TRUEthe nominated individuals that do not show inidvarare set toNA(see details).- timevar
Character sacalar. In the case of longitudinal data, name of the time var.
- t
Integer scalar. Repeat the network
ttimes (if not0,t1are provided).- undirected
Logical scalar. When
TRUEonly the lower triangle of the adjacency matrix will considered (faster).- self
Logical scalar. When
TRUEautolinks (loops, self edges) are allowed (see details).- multiple
Logical scalar. When
TRUEallows multiple edges.- keep.isolates
Logical scalar. When FALSE, rows with
NA/NULLvalues (isolated vertices unless have autolink) will be droped (see details).- recode.ids
Logical scalar. When TRUE ids are recoded using
as.factor(see details).- warn.coercion
Logical scalar. When
TRUEwarns coercion from numeric to integer.- ...
Further arguments to be passed to
new_diffnet.- edgelist
Two column matrix/data.frame in the form of ego -source- and alter -target- (see details).
- w
Numeric vector. Strength of ties (optional).
- t0
Integer vector. Starting time of the ties (optional).
- t1
Integer vector. Finishing time of the ties (optional).
- fill.missing
Character scalar. In the case of having unmatching ids between
datandedgelist, fills the data (see details).
Value
A diffnet object.
Details
All of netvars, toavar and groupvar
must be integers. Were these numeric they are coerced into integers, otherwise,
when neither of both, the function returns with error. idvar, on the
other hand, should only be integer when calling survey_to_diffnet,
on the contrary, for edgelist_to_diffnet, idvar may be character.
In field work it is not unusual that some respondents nominate unsurveyed
individuals. In such case, in order to exclude them from the analysis,
the user can set no.unsurveyed=TRUE (the default), telling the
function to exclude such individuals from the adjacency matrix. This is
done by setting variables in netvars equal to NA when the
nominated id can't be found in idvar.
If the network nomination process was done in different groups (location
for example) the survey id numbers may be define uniquely within each group
but not across groups (there may be many individuals with id=1,
for example). To encompass this issue, the user can tell the function what
variable can be used to distinguish between groups through the groupvar
argument. When groupvar is provided, function redifines idvar
and the variables in netvars as follows:
dat[[idvar]] <- dat[[idvar]] + dat[[groupvar]]*zWhere z = 10^nchar(max(dat[[idvar]])).
For longitudinal data, it is assumed that the toavar holds the same
information through time, this is, time-invariable. This as the package does
not yet support variable times of adoption.
The fill.missing option can take any of these three values: "edgelist",
"dat", or "both". This argument works as follows:
When
fill.missing="edgelist"(or"both") the function will check which vertices show indatbut do not show inedgelist. If there is any, the function will include these inedgelistas ego toNA(so they have no link to anyone), and, if specified, will fill thet0,t1vectors withNAs for those cases. Ifwis also specified, the new vertices will be set tomin(w, na.rm=TRUE).When
fill.missing="dat"(or"both") the function checks which vertices show inedgelistbut not indat. If there is any, the function will include these indatby adding one row per individual.
See also
Other data management functions:
diffnet-class,
edgelist_to_adjmat(),
egonet_attrs(),
isolated()
Examples
# Loading a fake survey (data frame)
data(fakesurvey)
# Diffnet object keeping isolated vertices ----------------------------------
dn1 <- survey_to_diffnet(fakesurvey, "id", c("net1", "net2", "net3"), "toa",
"group", keep.isolates=TRUE)
# Diffnet object NOT keeping isolated vertices
dn2 <- survey_to_diffnet(fakesurvey, "id", c("net1", "net2", "net3"), "toa",
"group", keep.isolates=FALSE)
# dn1 has an extra vertex than dn2
dn1
#> Dynamic network of class -diffnet-
#> Name : Diffusion Network
#> Behavior : Unspecified
#> # of nodes : 9 (101, 102, 103, 104, 105, 201, 202, 205, ...)
#> # of time periods : 5 (1 - 5)
#> Type : directed
#> Final prevalence : 0.89
#> Static attributes : group, net1, net2, net3, age, gender, note (7)
#> Dynamic attributes : -
dn2
#> Dynamic network of class -diffnet-
#> Name : Diffusion Network
#> Behavior : Unspecified
#> # of nodes : 8 (101, 102, 103, 104, 105, 201, 205, 210)
#> # of time periods : 5 (1 - 5)
#> Type : directed
#> Final prevalence : 0.88
#> Static attributes : group, net1, net2, net3, age, gender, note (7)
#> Dynamic attributes : -
# Loading a longitudinal survey data (two waves) ----------------------------
data(fakesurveyDyn)
groupvar <- "group"
x <- survey_to_diffnet(
fakesurveyDyn, "id", c("net1", "net2", "net3"), "toa", "group" ,
timevar = "time", keep.isolates = TRUE, warn.coercion=FALSE)
plot_diffnet(x, vertex.label = rownames(x))
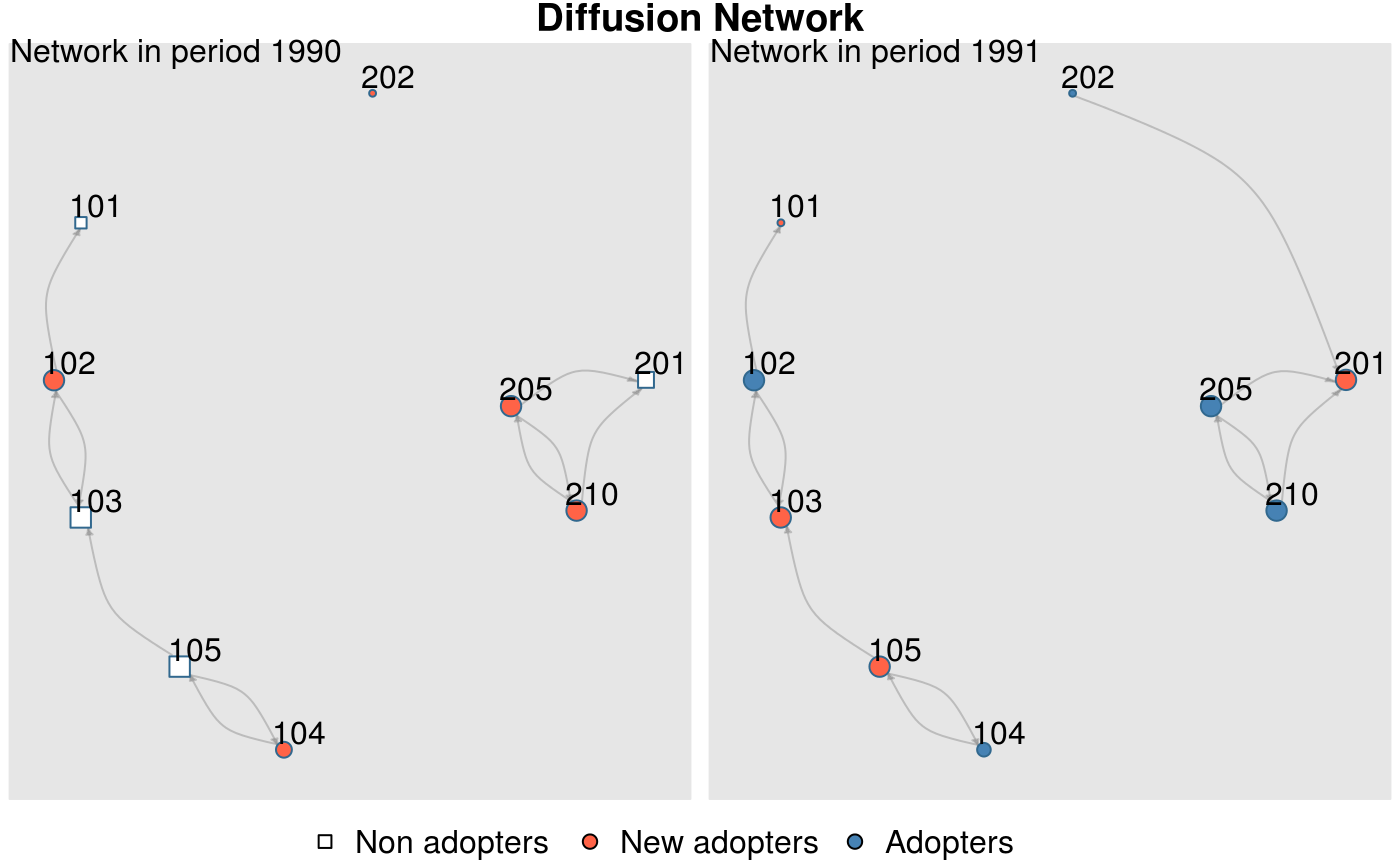 # Reproducing medInnovationsDiffNet object ----------------------------------
data(medInnovations)
# What are the netvars
netvars <- names(medInnovations)[grepl("^net", names(medInnovations))]
medInnovationsDiffNet2 <- survey_to_diffnet(
medInnovations,
"id", netvars, "toa", "city",
warn.coercion=FALSE)
medInnovationsDiffNet2
#> Dynamic network of class -diffnet-
#> Name : Diffusion Network
#> Behavior : Unspecified
#> # of nodes : 125 (1001, 1002, 1003, 1004, 1005, 1006, 1007, 1008, ...)
#> # of time periods : 18 (1 - 18)
#> Type : directed
#> Final prevalence : 1.00
#> Static attributes : city, detail, meet, coll, attend, proage, length, ... (58)
#> Dynamic attributes : -
# Comparing with the package's version
all(diffnet.toa(medInnovationsDiffNet2) == diffnet.toa(medInnovationsDiffNet)) #TRUE
#> [1] TRUE
all(
diffnet.attrs(medInnovationsDiffNet2, as.df = TRUE) ==
diffnet.attrs(medInnovationsDiffNet, as.df = TRUE),
na.rm=TRUE) #TRUE
#> [1] TRUE
# Reproducing medInnovationsDiffNet object ----------------------------------
data(medInnovations)
# What are the netvars
netvars <- names(medInnovations)[grepl("^net", names(medInnovations))]
medInnovationsDiffNet2 <- survey_to_diffnet(
medInnovations,
"id", netvars, "toa", "city",
warn.coercion=FALSE)
medInnovationsDiffNet2
#> Dynamic network of class -diffnet-
#> Name : Diffusion Network
#> Behavior : Unspecified
#> # of nodes : 125 (1001, 1002, 1003, 1004, 1005, 1006, 1007, 1008, ...)
#> # of time periods : 18 (1 - 18)
#> Type : directed
#> Final prevalence : 1.00
#> Static attributes : city, detail, meet, coll, attend, proage, length, ... (58)
#> Dynamic attributes : -
# Comparing with the package's version
all(diffnet.toa(medInnovationsDiffNet2) == diffnet.toa(medInnovationsDiffNet)) #TRUE
#> [1] TRUE
all(
diffnet.attrs(medInnovationsDiffNet2, as.df = TRUE) ==
diffnet.attrs(medInnovationsDiffNet, as.df = TRUE),
na.rm=TRUE) #TRUE
#> [1] TRUE