# Setting the seed for the RNG
set.seed(1213)
# Generating a random diffusion network
net <- rdiffnet(
n = 1e3, # 1.
t = 20, # 2.
seed.nodes = "random", # 3.
seed.p.adopt = .1, # 4.
seed.graph = "small-world", # 5.
rgraph.args = list(p=.2), # 6.
threshold.dist = function(x) runif(1, .3, .7) # 7.
)Simulation of diffusion networks: rdiffnet
Recap
Network thresholds (Valente, 1995; 1996), \(\tau\), are defined as the required proportion or number of neighbors that leads you to adopt a particular behavior (innovation), \(a=1\). So, in simulations, the adoption rule follows
\[ a_i = \left\{\begin{array}{ll} 1 &\mbox{if } \tau_i\leq E_i \\ 0 & \mbox{Otherwise} \end{array}\right. ,\qquad \text{where} \quad E_i \equiv \frac{\sum_{j\neq i}\mathbf{X}_{ij}a_j}{\sum_{j\neq i}\mathbf{X}_{ij}}. \]
Where \(E_i\) is i’s exposure to the innovation and \(\mathbf{X}\) is the adjacency matrix (the network).
Here is a review of the concepts we will be using:
- Exposure \(E_i\) : Proportion/number of neighbors that has adopted an innovation at each point in time.
- Threshold \(\tau\) : The proportion/number of your neighbors who had adopted at or one time period before ego (the focal individual) adopted.
- Infectiousness: How much \(i\)’s adoption affects her alters.
- Susceptibility: How much \(i\)’s alters’ adoption affects her.
- Structural equivalence: How similar are \(i\) and \(j\) in terms of position in the network.
Simulating diffusion networks
We will simulate a diffusion network with the following parameters, using the rdiffnet function included in the package::
- Will have 1,000 vertices,
- Will span 20 time periods,
- The initial adopters (seeds) will be selected random,
- Seeds will be a 10% of the network,
- The graph (network) will be small-world,
- Will use the WS algorithmwith \(p=.2\) (probability of rewire).
- Threshold levels will be uniformly distributed between [0.3, 0.7]
Main features of
rdiffnet:Generatting complex graphs or using your own,
Setting threshold levels per node,
Network rewiring throughout the simulation, and
Setting the seed nodes.
The simulation algorithm is as follows:
If required, a baseline graph is created,
Set of initial adopters and threshold distribution are established,
The set of t networks is created (if required), and
Simulation starts at t=2, assigning adopters based on exposures and thresholds:
For each \(i \in N\), if its exposure at \(t-1\) is greater than its threshold, then adopts, otherwise continue without change.
next \(i\)
Rumor spreading vs Disease
library(netdiffuseR)
set.seed(09)
diffnet_disease <- rdiffnet(
n = 5e2,
t = 5,
seed.graph = "small-world",
rgraph.args = list(k = 4, p = .3),
seed.nodes = "random",
seed.p.adopt = .05,
rewire = TRUE,
threshold.dist = function(i) 1L,
exposure.args = list(normalized = FALSE),
name = "Disease spreading"
)
diffnet_rumor <- rdiffnet(
seed.graph = diffnet_disease$graph,
seed.nodes = which(diffnet_disease$toa == 1),
rewire = FALSE,
threshold.dist = function(i) rbeta(1, 3, 10),
name = "Rumor spreading",
behavior = "Some rumor"
)
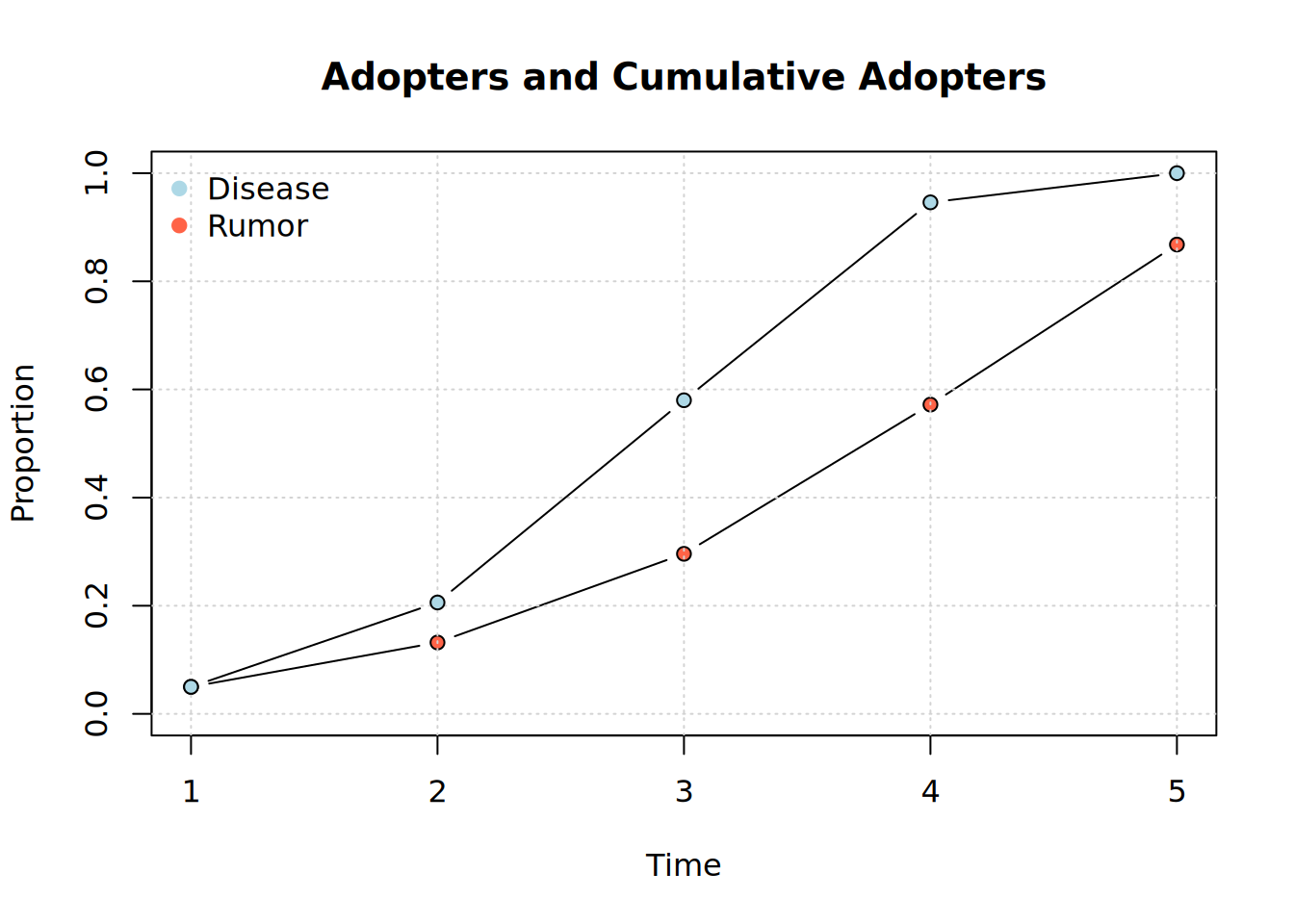
plot_adopters(diffnet_rumor, what = "cumadopt", include.legend = FALSE)
plot_adopters(diffnet_disease, bg="lightblue", add=TRUE, what = "cumadopt")
legend(
"topleft",
legend = c("Disease", "Rumor"),
col = c("lightblue", "tomato"),
bty = "n", pch=19
)
Problems
Using the Korean family village 21, compare which strategy setting the seed nodes maximizes the diffusion. Use
rdiffnet_multipleto run 500 simulations for each strategy. The strategies are:- Random seed nodes.
- Central seed nodes.
- Marginal seed nodes.
- One of your own making.
(solution script and solution plot)
- Given the following types of networks: Small-world, Scale-free, Bernoulli, what set of \(n\) initiators maximizes diffusion? (solution script and solution plot)
Appendix
Not using rdiffnet_multiple
The following is example code that can be used to run multiple simulations like it is done using the rdiffnet_multiple function. We do not recommend this approach but it may be useful for some users:
# Now, simulating a bunch of diffusion processes
nsim <- 500L
ans_1and2 <- vector("list", nsim)
set.seed(223)
for (i in 1:nsim) {
# We just want the cum adopt count
ans_1and2[[i]] <-
cumulative_adopt_count(
rdiffnet(
seed.graph = net,
threshold.dist = sample(1:2, 1000L, TRUE),
seed.nodes = "random",
seed.p.adopt = .10,
exposure.args = list(outgoing = FALSE, normalized = FALSE),
rewire = FALSE
)
)
# Are we there yet?
if (!(i %% 50))
message("Simulation ", i," of ", nsim, " done.")
}# Simulation 50 of 500 done.# Simulation 100 of 500 done.# Simulation 150 of 500 done.# Simulation 200 of 500 done.# Simulation 250 of 500 done.# Simulation 300 of 500 done.# Simulation 350 of 500 done.# Simulation 400 of 500 done.# Simulation 450 of 500 done.# Simulation 500 of 500 done.# Extracting prop
ans_1and2 <- do.call(rbind, lapply(ans_1and2, "[", i="prop", j=))
ans_2and3 <- vector("list", nsim)
set.seed(223)
for (i in 1:nsim) {
# We just want the cum adopt count
ans_2and3[[i]] <-
cumulative_adopt_count(
rdiffnet(
seed.graph = net,
threshold.dist = sample(2:3, 1000L, TRUE),
seed.nodes = "random",
seed.p.adopt = .10,
exposure.args = list(outgoing = FALSE, normalized = FALSE),
rewire = FALSE
)
)
# Are we there yet?
if (!(i %% 50))
message("Simulation ", i," of ", nsim, " done.")
}# Simulation 50 of 500 done.# Simulation 100 of 500 done.# Simulation 150 of 500 done.# Simulation 200 of 500 done.# Simulation 250 of 500 done.# Simulation 300 of 500 done.# Simulation 350 of 500 done.# Simulation 400 of 500 done.# Simulation 450 of 500 done.# Simulation 500 of 500 done.ans_2and3 <- do.call(rbind, lapply(ans_2and3, "[", i="prop", j=))Example by changing threshold (rdiffnet_multiple)
The following block of code runs multiple diffnet simulations.
Before we proceed, we will generate a scale-free homophilic network using rgraph_ba:
# Simulating a scale-free homophilic network
set.seed(1231)
X <- rep(c(1,1,1,1,1,0,0,0,0,0), 50)
net <- rgraph_ba(t = 499, m=4, eta = X)
# Taking a look in igraph
ig <- igraph::graph_from_adjacency_matrix(net)
plot(ig, vertex.color = c("azure", "tomato")[X+1], vertex.label = NA,
vertex.size = sqrt(dgr(net)))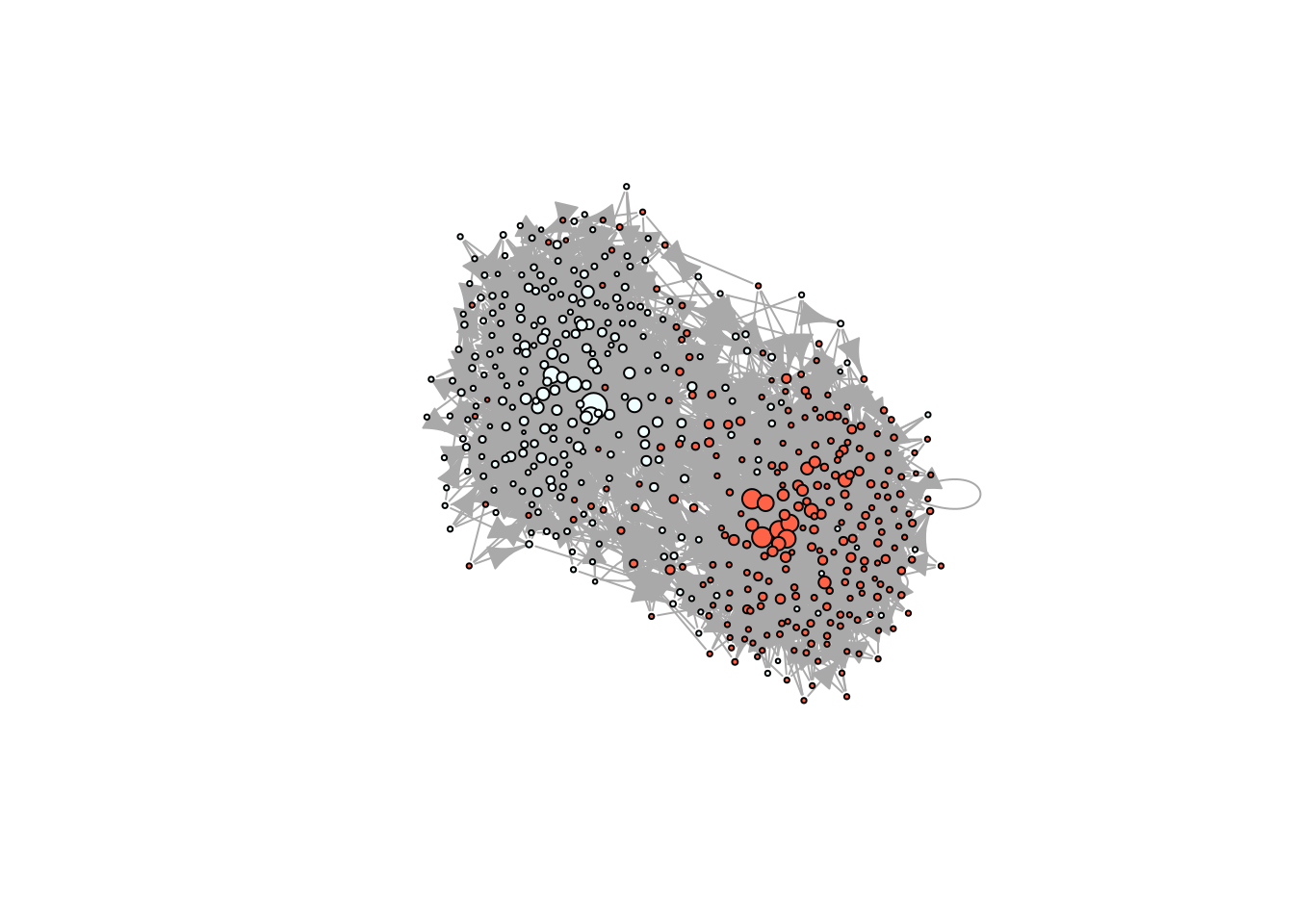
Besides of the usual parameters passed to rdiffnet, the rdiffnet_multiple function requires:
R(number of repetitions/simulations), andstatistic(a function that returns the statistic of insterst).
Optionally, the user may choose to specify the number of clusters to run it in parallel (multiple CPUs):
nsim <- 500L
ans_1and2 <- rdiffnet_multiple(
# Num of sim
R = nsim,
# Statistic
statistic = function(d) cumulative_adopt_count(d)["prop",],
seed.graph = net,
t = 10,
threshold.dist = sample(1:2, 500L, TRUE),
seed.nodes = "random",
seed.p.adopt = .1,
rewire = FALSE,
exposure.args = list(outgoing=FALSE, normalized=FALSE),
# Running on 4 cores
ncpus = 4L
) |> t()
ans_2and3 <- rdiffnet_multiple(
# Num of sim
R = nsim,
# Statistic
statistic = function(d) cumulative_adopt_count(d)["prop",],
seed.graph = net,
t = 10,
threshold.dist = sample(2:3, 500, TRUE),
seed.nodes = "random",
seed.p.adopt = .1,
rewire = FALSE,
exposure.args = list(outgoing=FALSE, normalized=FALSE),
# Running on 4 cores
ncpus = 4L
) |> t()
ans_1and3 <- rdiffnet_multiple(
# Num of sim
R = nsim,
# Statistic
statistic = function(d) cumulative_adopt_count(d)["prop",],
seed.graph = net,
t = 10,
threshold.dist = sample(1:3, 500, TRUE),
seed.nodes = "random",
seed.p.adopt = .1,
rewire = FALSE,
exposure.args = list(outgoing=FALSE, normalized=FALSE),
# Running on 4 cores
ncpus = 4L
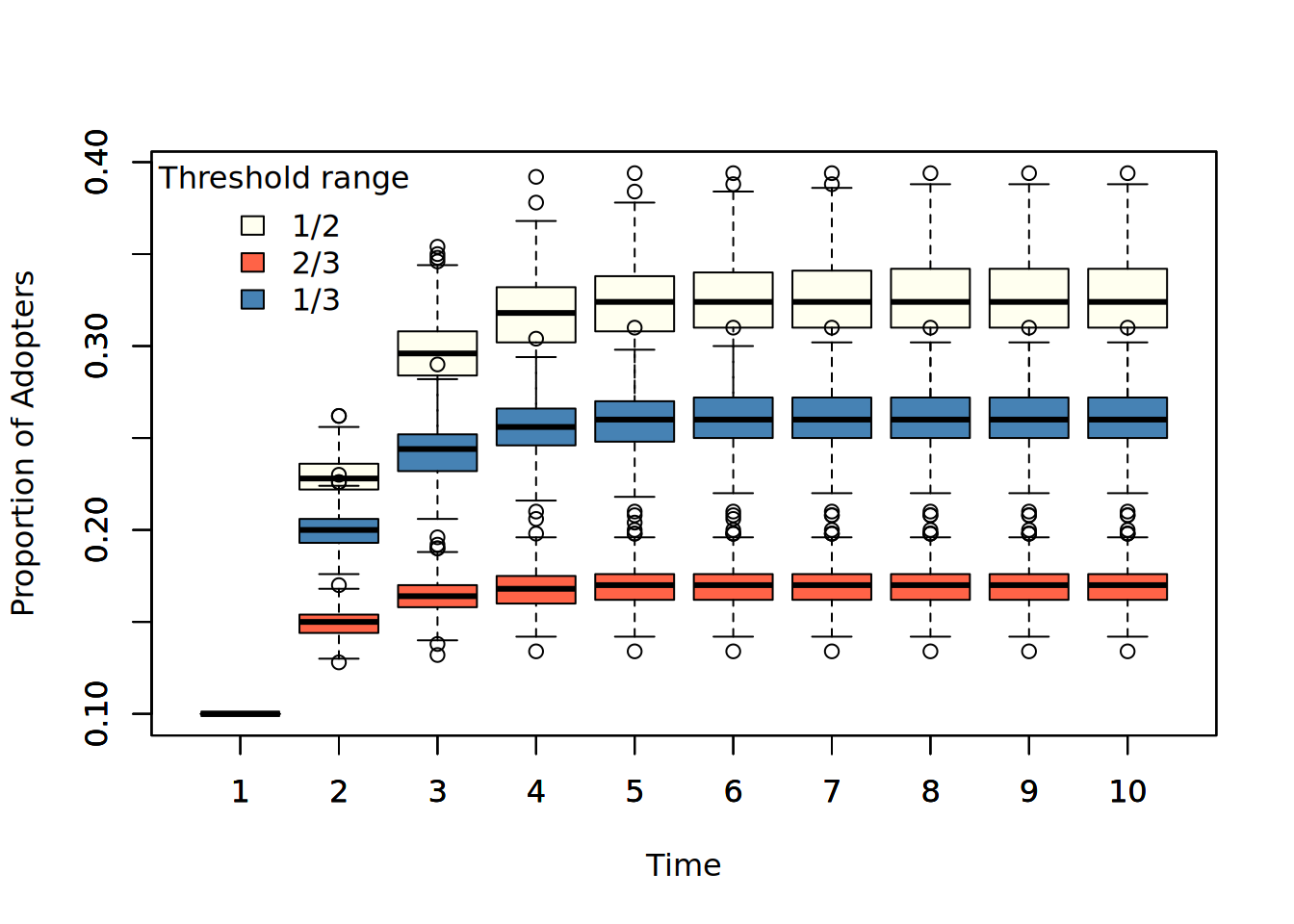
) |> t()- By simulating 1000 times each diffusion, we can see the final prevalence is a function of threshold levels.
boxplot(ans_1and2, col="ivory", xlab = "Time", ylab = "Proportion of Adopters")
boxplot(ans_2and3, col="tomato", add=TRUE)
boxplot(ans_1and3, col = "steelblue", add=TRUE)
legend(
"topleft",
fill = c("ivory", "tomato", "steelblue"),
legend = c("1/2", "2/3", "1/3"),
title = "Threshold range",
bty ="n"
)