Computes the requested degree measure for each node in the graph.
dgr(
graph,
cmode = "degree",
undirected = getOption("diffnet.undirected", FALSE),
self = getOption("diffnet.self", FALSE),
valued = getOption("diffnet.valued", FALSE)
)
# S3 method for class 'diffnet_degSeq'
plot(
x,
breaks = min(100L, nrow(x)/5),
freq = FALSE,
y = NULL,
log = "xy",
hist.args = list(),
slice = ncol(x),
xlab = "Degree",
ylab = "Freq",
...
)Arguments
- graph
Any class of accepted graph format (see
netdiffuseR-graphs).- cmode
Character scalar. Either "indegree", "outdegree" or "degree".
- undirected
Logical scalar. When
TRUEonly the lower triangle of the adjacency matrix will considered (faster).- self
Logical scalar. When
TRUEautolinks (loops, self edges) are allowed (see details).- valued
Logical scalar. When
TRUEweights will be considered. Otherwise non-zero values will be replaced by ones.- x
An
diffnet_degSeq object- breaks
Passed to
hist.- freq
Logical scalar. When
TRUEthe y-axis will reflex counts, otherwise densities.- y
Ignored
- log
- hist.args
Arguments passed to
hist.- slice
Integer scalar. In the case of dynamic graphs, number of time point to plot.
- xlab
Character scalar. Passed to
plot.- ylab
Character scalar. Passed to
plot.- ...
Further arguments passed to
plot.
Value
A numeric matrix of size \(n\times T\). In the case of plot,
returns an object of class histogram.
See also
Other statistics:
bass,
classify_adopters(),
cumulative_adopt_count(),
ego_variance(),
exposure(),
hazard_rate(),
infection(),
moran(),
struct_equiv(),
threshold(),
vertex_covariate_dist()
Other visualizations:
diffusionMap(),
drawColorKey(),
grid_distribution(),
hazard_rate(),
plot_adopters(),
plot_diffnet2(),
plot_diffnet(),
plot_infectsuscep(),
plot_threshold(),
rescale_vertex_igraph()
Examples
# Comparing degree measurements ---------------------------------------------
# Creating an undirected graph
graph <- rgraph_ba()
graph
#> 11 x 11 sparse Matrix of class "dgCMatrix"
#> [[ suppressing 11 column names ‘1’, ‘2’, ‘3’ ... ]]
#>
#> 1 1 . . . . . . . . . .
#> 2 1 . . . . . . . . . .
#> 3 1 . . . . . . . . . .
#> 4 . 1 . . . . . . . . .
#> 5 1 . . . . . . . . . .
#> 6 . . . . . 1 . . . . .
#> 7 . . . . . 1 . . . . .
#> 8 . 1 . . . . . . . . .
#> 9 . . . . . . . 1 . . .
#> 10 . . . . 1 . . . . . .
#> 11 . . . . 1 . . . . . .
data.frame(
In=dgr(graph, "indegree", undirected = FALSE),
Out=dgr(graph, "outdegree", undirected = FALSE),
Degree=dgr(graph, "degree", undirected = FALSE)
)
#> In Out Degree
#> 1 3 0 3
#> 2 2 1 3
#> 3 0 1 1
#> 4 0 1 1
#> 5 2 1 3
#> 6 1 0 1
#> 7 0 1 1
#> 8 1 1 2
#> 9 0 1 1
#> 10 0 1 1
#> 11 0 1 1
# Testing on Korean Family Planning (weighted graph) ------------------------
data(kfamilyDiffNet)
d_unvalued <- dgr(kfamilyDiffNet, valued=FALSE)
d_valued <- dgr(kfamilyDiffNet, valued=TRUE)
any(d_valued!=d_unvalued)
#> [1] TRUE
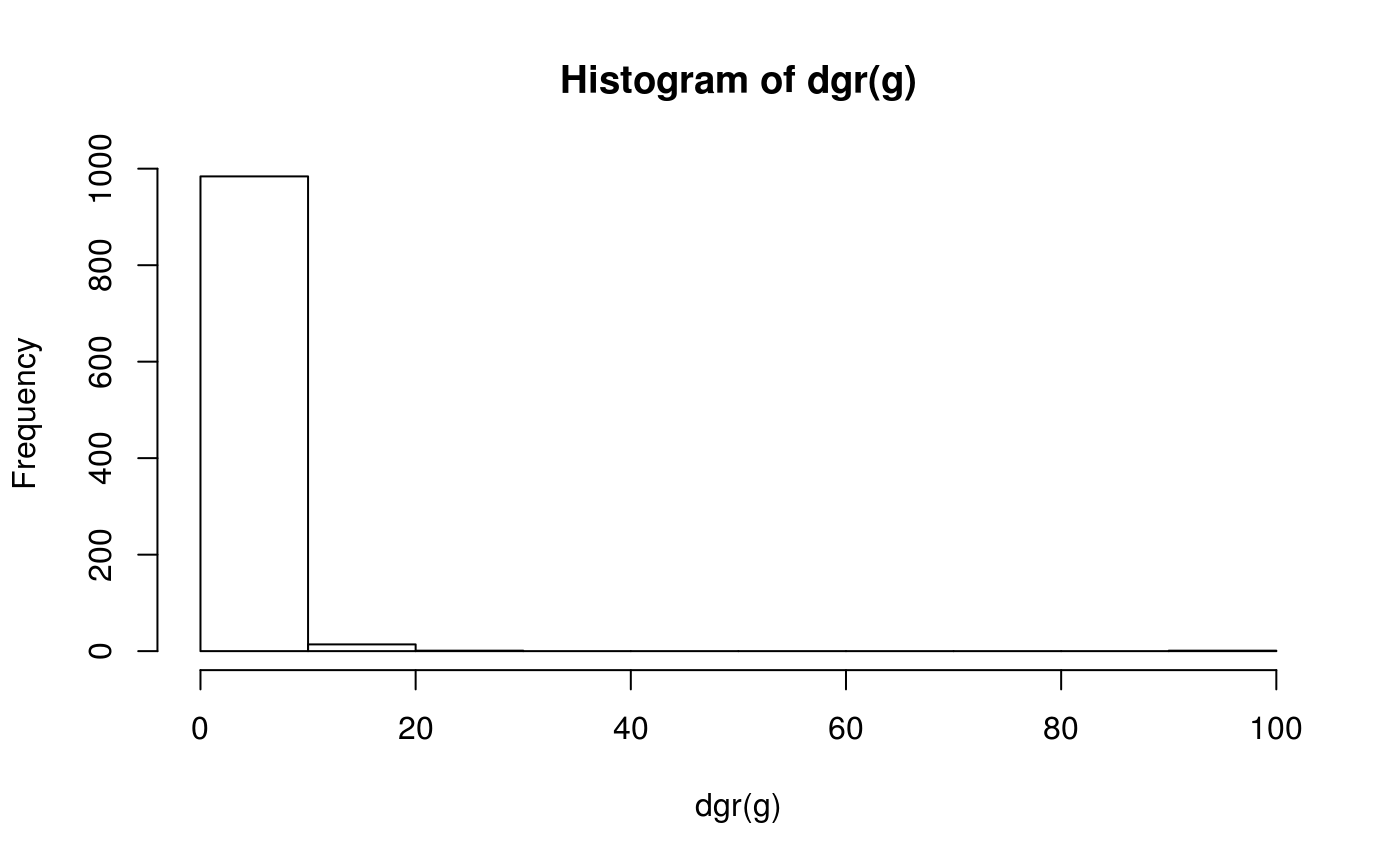
# Classic Scale-free plot ---------------------------------------------------
set.seed(1122)
g <- rgraph_ba(t=1e3-1)
hist(dgr(g))
 # Since by default uses logscale, here we suppress the warnings
# on points been discarded for <=0.
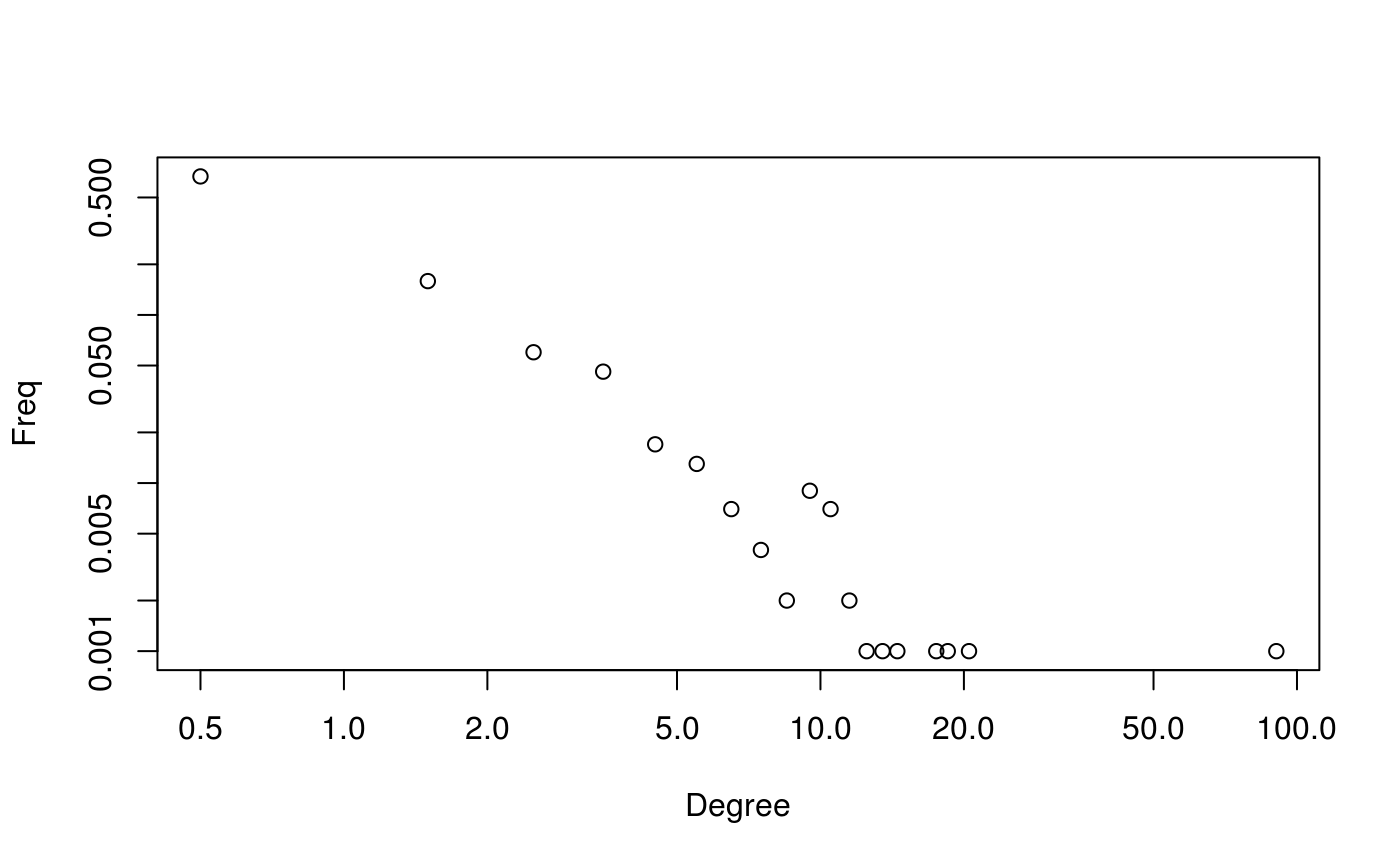
suppressWarnings(plot(dgr(g)))
# Since by default uses logscale, here we suppress the warnings
# on points been discarded for <=0.
suppressWarnings(plot(dgr(g)))